Bioremediation Potential of Bacterial Isolates for Municipal Wastewater Treatment
Nilesh A. Sonune1 * and Anil M. Garode2
1
School of Life Sciences,
Swami Ramanand Teerth Marathwada University,
Nanded,
431606
Maharashtra
India
2
Department of Microbiology,
District Buldana,
Shri Shivaji Science College Chikhli,
443201
Maharashtra
India
Corresponding author Email: nasonune@gmail.com
DOI: http://dx.doi.org/10.12944/CWE.10.2.27
The potential of bacteria for the treatment of municipal wastewater was investigated in present study. Total eight bacterial isolates were used for this study that showed growth on wastewater agar medium. These isolates were identified on the basis of morphological and biochemical test and identified as Bacillus licheniformis NW16, Pseudomonas aeruginosa NS19, Pseudomonas sp. NS20, Planococcus salinarum NS23, Stenotrophomonas maltophilia NS21, Paenibacillus sp. NW9, Paenibacillus borealis NS3 and Aeromonas hydrophilia NS17. The B. licheniformis NW16 showed highest potential to reduce all parameter under study than other isolates except Ammonical nitrogen. B. licheniformis NW16 and Aeromonas hydrophilia NS17 showed maximum reduction (42.86%) in BOD each. B. licheniformis NW16 and Paenibacillus sp. NW9 showed 82.76% and 81.61% reduction in COD respectively. B. licheniformis NW16, P. salinarum NS23 and Aeromonas hydrophilia NS17 showed reduction in nitrate ranging from 17.36%-63.64%. All the isolates have potential to reduced phosphate from 17.55% -72.3%. B. licheniformis NW16, Ps. aeruginosa NS19, Pseudomonas sp. NS20, Paenibacillus sp. NW9 and Aeromonas hydrophilia NS17 showed reduction in TSS ranging from 42.69%-79.94%. B. licheniformis NW16, Ps. aeruginosa NS19, Pseudomonas sp. NS20, S. maltophilia NS21 and Paenibacillus sp. NW9 showed reduction in TDS ranging from 14%-81.4%.
Copy the following to cite this article:
Sonune N. A, Garode A. M. Bioremediation Potential of Bacterial Isolates for Municipal Wastewater Treatment. Curr World Environ 2015;10(2) DOI:http://dx.doi.org/10.12944/CWE.10.2.27
Copy the following to cite this URL:
Sonune N. A, Garode A. M. Bioremediation Potential of Bacterial Isolates for Municipal Wastewater Treatment. Curr World Environ 2015;10(2). Available from: http://www.cwejournal.org/?p=11902
Download article (pdf) Citation Manager Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2015-06-16 |
|---|---|
| Accepted: | 2015-07-19 |
Introduction
Water is one of the most important natural resource required to all living organisms. Its diversified uses include drinking, cooking, washing, irrigation and industrial activities (Rathore et al., 2014). Recently, water pollution is main problem because of uncontrolled urbanization which is due to sewage effluent disposed into water bodies and leads to the adverse effect on living organism (Tamil Selvi et al., 2012). Due to such problems the main global agenda is environmental management, treatment and disposal, wastes recycling, pollution control and prevention and reuse of the wastewater (Azab, 2008).
Sewage water is a complex matrix. These include high concentration of BOD, COD and high dissolved solid. The quality of wastewater is determined by analysing parameters such as COD, BOD, nitrate, TSS, TDS etc. These parameters provide crucial information of the quality of the wastewater (Loos et al., 2013). Wastewater is generated by residential, institutional, commercial and industrial establishments and includes household liquid wastes from baths, toilets, kitchens and sinks that are disposed off via sewers. The composition of sewage is differ widely but may contain more than 95% water with pathogens i.e. bacteria, viruses and parasitic worms and non-pathogenic bacteria. Chemical contaminants include organic particles, inorganics particles (Shannon et al., 2007).
Global attention has been drawn on ways to sustain the environment using microorganism to remediate environmental pollutants because physical and chemical treatment are costly and can lead to production of toxic substance (Luka et al., 2014). Bioremediation involves the use of microorganism to reduce or remove the pollutants from contaminated area which may lead to restoration of the original natural substance without further disruption to the local environment (Vidali, 2001; Deveraja et al., 2002; Vezzulli et al., 2004). Mostly, the oxidized products of organic materials are CO2 and new microbial cells. The organic matter provides energy and carbon as a nutrient source for cell growth (Chui et al., 2006). Bioremediation is an economical, eco-friendly and requires less expensive techniques for water pollution. But the correct microbe should be utilized in the appropriate place with the precise environmental factors (Boopathy, 2000). Therefore, the main goal of present study was to examine the ability of indigenous bacteria for bioremediation of the municipal wastewater.
Materials And Methods
Sample collection and site
Wastewater and sludge samples were collected from various places from Buldana district, India, in pre-sterilized bottle and Zip-lock plastic bag respectively according to standard procedures from American Public Health Association (APHA, 2005) and transferred immediately to the laboratory.
Isolation and identification of bacterial isolates
Wastewater and sludge samples were serially diluted and inoculated on the Nutrient agar medium separately. Morphologically different colonies were isolated and maintained at 40C on nutrient agar slants. The purified isolates were identified by morphological and biochemical characteristics based on Bergey’s Manual of Determinative Bacteriology (Holt, 1994).
Preliminary screening of efficient bacterial isolates for bioremediation study
All bacterial isolates were inoculated on wastewater agar medium (WWA). The composition of the medium per 100 ml was 100 ml sterilized wastewater and 2% agar. All plates were incubated for 48 hr at 370C. Those bacterial isolates which showed growth on WWA medium were used for bioremediation studies.
Characterization of wastewater samples
Wastewater samples were characterized before and after the treatment. The parameters under study include pH, BOD, COD, Ammonical Nitrogen, Nitrate, Phosphate, TSS and TDS. All parameter were analyzed by using Standard Methods for the Examination of Water and Wastewater (APHA, 2005). Removal efficiencies of all parameters were calculated according to the following equation:
Removal Efficiency (RE %) = C0-RC/C0× 100
Where, C0=Initial Concentration before Treatment,
RC= Final Concentration after Treatment
Batch culture study of bacterial isolates
Each bacterial cultures were inoculated individually in pre-sterilized 50 ml wastewater broth (WWB) i,e. only wastewater, in this 0.5% peptone were added to enhance the growth of bacterial isolates. The flask was kept in a shaker at 120 rpm for 48 h at 370C. The O.D. of cell suspension was adjusted to 0.5 by using sterile saline solution (0.85%) at 600 nm. Inoculum of each isolates (10%) was taken in 250 ml flask containing 90 ml non-sterile wastewater separately. Along with this, one control flask was used containing only non-sterilized wastewater. These flasks were kept in shaker at 120 rpm for 72 hr. After treatment, all samples were centrifuged at 10,000 rpm for 20 minutes at 100C and supernatants were used for further analysis.
Results and Discussion
Isolation and characterization of bacterial isolates
Total eight bacterial isolates were identified on the basis of morphological and biochemical characteristics. It includes Bacillus licheniformis NW16, Ps. aeruginosa NS19, Pseudomonas sp. NS20, P. salinarum NS23, S. maltophilia NS21, Paenibacillus borealis NS3, Paenibacillus sp. NW9 and Aeromonas hydrophilia NS17.
Batch culture study of bacterial isolates
Total 44 bacterial isolates were isolated on nutrient agar medium. Out of these, 8 bacterial isolates showed growth on WWA medium. The organic matter contained in the wastewater provides a substrate for these isolates. These isolates were used for bioremediation studies by batch experiments.
The initial pH of the sample was slightly acidic whereas it was slightly alkaline after treatment. The control showed negligible change in pH (data is not shown here). Degradation of proteins and amino acids present in wastewater was converted into ammonia that increases the pH of sample to slightly alkaline. The change in pH of wastewater suggests that there has been activity of microorganisms which degrade organic matter.
Biochemical oxygen demand (BOD) is a measure of the oxygen required to microorganisms for the decomposition of waste material. The initial BOD of the samples was higher than the permissible limit. High BOD indicates high amount of organic matter, it lead to oxygen depletion and creates anaerobic conditions which would result in reduction of diversity and distribution of aquatic fauna. Organic matter will support anaerobic action leading to the accumulation of toxic compounds in water bodies (Goel, 1997). The maximum percentage removal of BOD in 72 hours was observed by B. licheniformis NW16 and Aeromonas hydrophilia NS17 i.e. 42.86% each followed by P. salinarum NS23, Ps. aeruginosa NS19, S. maltophilia NS21, Pseudomonas sp. NS20, Paenibacillus borealis NS3 and Paenibacillus sp. NW9 with 28.91%, 28.57%, 25.46%, 21.96%, 19.85% and 14.29% respectively whereas control showed only 8.93% (Fig. 1). Similar results were observed by Shrivastava et al., (2013) and Prasad and Manjunath (2011) and found that B. subtilis and Pseudomonas aeruginosa has BOD and COD reduction potential while studying on bioremediation of Yamuna water and lipid rich wastewater respectively. Vasconcellos et al., (2009) reported B. licheniformis for biodegradation of cassava processing wastewater whereas Ravi Kumar et al., (2013) reported 36.41% BOD removal efficiency by B. licheniformis for bioremediation of sewage.
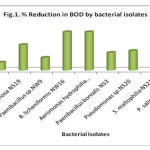 |
Figure 1: % Reduction in BOD by bacterial isolates. Click here to View figure |
Chemical Oxygen Demand (COD) test is the best and rapid method for estimation of organic matter present in the wastewater sample. In our study it was observed that all isolates showing reduction in COD after 72 hr (Fig. 2). B. licheniformis NW16 showed maximum reduction in COD (82.76%) followed by Paenibacillus sp. NW9, Pseudomonas sp. NS20, Aeromonas hydrophilia NS17, Paenibacillus borealis NS3, P. salinarum NS23, Ps. aeruginosa NS19 and S. maltophilia NS21 with 81.61%, 60%, 55.3%, 30%, 22.5%, 21% and 20% respectively and compared with control which showed 14.94% reduction in COD. In biodegradation, bacteria uses organic compound as a substrate for their growth and development (Chin et al., 1995). These bacteria are capable of producing a wide variety of enzymes that can degrade complex organic compounds into CO2 and water present in the wastewater (Claxton and Houx, 1995; Chin et al., 1995). The bacterial species present in the wastewater has no significant effect on removal of BOD and COD as observed in case of control. However, our isolates showed promising results. Similar results were observed by Gaikwad et al., (2014) and Zhao et al., (2009) found that Pseudomonas sp. and Bacillus sp. were able to reduced COD and BOD. Mazzucotelli et al., (2014) reported use of Stenotrophomonas for degradation of dairy wastewater and Ji-hong et al., (2008) also reported COD reducer Aeromonas sp.
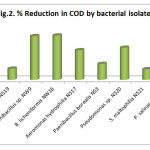 |
Figure 2: % Reduction in COD by bacterial isolates Click here to View figure |
In the present study, none of the bacterial isolates showed reduction in Ammonical nitrogen (Fig. 3). The increase in the concentration of Ammonical nitrogen was because of degradation of protein and nitrogenous compounds into ammonia.
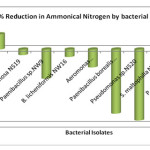 |
Figure 3: % Reduction in Ammonical Nitrogen by bacterial isolates Click here to View figure |
The nitrate concentration prior to treatment was very high than permissible limits. Nitrate is one of the source for eutrophication of water. In the present study, the maximum reduction in nitrate was showed by B. licheniformis NW16 (63.64%), P. salinarum NS23 (27.87%) and Aeromonas hydrophilia NS17 (17.37%). It was observed that the increase in the concentration of nitrate in case of Ps. aeruginosa NS19, Pseudomonas sp. NS20, S. maltophilia NS21, Paenibacillus borealis NS3 and Paenibacillus sp. NW9 suggests the process of nitrification whereas decrease in nitrate concentration in case of B. licheniformis NW16, P. salinarum NS23, Aeromonas hydrophilia NS17 showed denitrification process that is conversion of nitrate into the molecular nitrogen (Fig. 4). Rajakumar et al., (2008) reported that Bacillus sp. and Pseudomonas sp. were most efficient for nitrate reduction. In our study, Bacillus sp. showed reduction in nitrate whereas Pseudomonas sp. showed contrasting result.
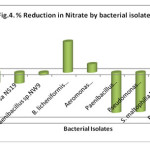 |
Figure 4: % Reduction in Nitrate by bacterial isolates Click here to View figure |
The phosphate is one of the most serious environmental problems because of its contribution to the eutrophication process of lakes and other natural waters. It occurs in natural water, wastewater, sediments and sludge. The possible entry of this ion into aquatic environment is through household sewage. The reduction showed by Paenibacillus borealis NS3 (72.3%), Ps. aeruginosa NS19 and B. licheniformis NW16 (59.98%) each, Paenibacillus sp. NW9, (55.06%), Pseudomonas sp. NS20 (47.69%), S. maltophilia NS21 (41.54%), P. salinarum NS23 (24.61%) and Aeromonas hydrophilia NS17 (17.55%). The control showed no removal in phosphate concentration (Fig. 5). Similar results were observed by Krishnaswamy et al., (2011) and found that the Bacillus sp RS-1 and Pseudomonas sp. YLW-7 were found to be efficient in phosphate reduction.
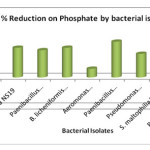 |
Figure 5: % Reduction on Phosphate by bacterial isolates Click here to View figure |
High amount of suspended particles has detrimental effects on aquatic flora and fauna and reduce the diversity of life in aquatic system and promote depletion of oxygen (Karthikeyan et al., 2010). The results from Fig.6 suggested that the maximum reduction in TSS was showed by Aeromonas hydrophilia NS17 (79.94%) followed by B. licheniformis NW16 (67.88%), Ps. Aeruginosa NS19 (57.88%), Paenibacillus sp. NW9 (54.15%), Pseudomonas sp. NS20 (42.69%) whereas Paenibacillus borealis NS3, P. salinarum NS23, S. maltophilia NS21 and control were unable to reduce TSS.
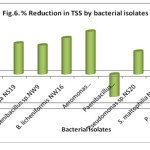 |
Figure 6: % Reduction in TSS by bacterial isolates Click here to View figure |
Total dissolved solid refers to all dissolved materials present in the wastewater. Discharge of wastewater with a high TDS level would have adverse impact on aquatic life and reduces crop yields if used for irrigation. In this study most of the isolates showed reduction in TDS. The maximum reduction in TDS was showed by Paenibacillus sp. NW9 (81.4 %,), S. maltophilia NS21 (76.74%), B. licheniformis NW16 (71.08%), Ps. aeruginosa NS19 (68.6%), Aeromonas hydrophilia NS17 (62.79%), control (32%), Pseudomonas sp. NS20 (6.98%) whereas Paenibacillus borealis NS3 and P. salinarum NS23 were unable to reduce TDS (Fig. 7). Tomar and Mittal (2014) also found that B. subtilis has potential for reduction of TSS and TDS.
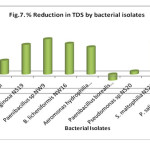 |
Figure 7: % Reduction in TDS by bacterial isolates Click here to View figure |
Conclusion
In the present study, total eight bacterial isolates namely B. licheniformis NW16, Ps. Aeruginosa NS19, Pseudomonas sp. NS20, P. salinarum NS23, S. maltophilia NS21, Paenibacillus borealis NS3, Paenibacillus sp. NW9 and Aeromonas hydrophilia NS17 were isolated and used for bioremediation of municipal wastewater. These isolates were showed degradation of organic matter in term of BOD, COD, nitrate, phosphate, TSS and TDS. Hence, these isolates may be used for bioremediation of municipal wastewater to control water pollution.
References
- Rathore DS, Rai N, Ashiya P, Physico Chemical Analysis of Water of Ayad River at Udaipur, Rajasthan (India). International Journal of Innovative Research in Science, Engineering and Technology 3(4): 11660- 11667 (2014).
- Tamil selvi A, Anjugam E, Archana Devi R, Madhan B, Kannappan S, Isolation and characterization of Bacteria from tannery Effluent Treatment plant and their Tolerance to Heavy metals and antibodies. Asian J Exp Biol Sci 3: 34-41 (2012).
- Azab MS, Waste-waste treatment technology and environmental management using sawdust bio-mixture. JTUSCI 1: 12-23 (2008).
- Loos R, Carvalho R, Antonio DC, Comero S, Locoro G, Tavazzi S, Paracchini B, Ghiani M, Lettieri T, Blaha L, Jarosova B, Voorspoels S, Servaes K, Haglund P, Fick J, Lindberg RH, EU-wide monitoring survey on emerging polar organic contaminants in wastewater treatment plant effluents. Water Research 47(17): 6475-6487 (2013).
- Shannon KE, Lee DY, Trevors JT, Beaudette LA, Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci Total Environ 382: 121-129 (2007).
- Luka Y, Usman FO, Tya TSK, Joseph C, Kinetics of Bioremediation of Lake Gerio in Jimeta-Yola Using Pseudomonas aerigunosa. International Refereed Journal of Engineering and Science 3 (2): 54-59 (2014).
- Vidali M, Bioremediation: An overview, Journal of Pure and Applied Chemistry 73 (7): 1163-1172 (2001).
- Devaraja TN, Yusoff FM, Shariff M, Changes in bacterial population and shrimp production in ponds treated with commercial microbial products. Aquaculture 206: 245–256 (2002).
- Vezzulli L, Pruzzo C, Fabiano M, Response of the bacterial community to in situ bioremediation of organic-rich sediments. Mar Pollut Bull 49: 740–751 (2004).
- Chui CH, Hau DK, Lau FY, Cheng GY, Wong RS, Gambari R, Apoptotic potential of the concentrated effective microorganism fermentation extract on human cancer cells. Int J Molecular Med 17(2): 279-284 (2006).
- Boopathy R, Formation of aniline as a transient metabolite during the metabolism of tetrl by a sulfate-reducing bacterial consortium. Current Microbiology 40: 190-193 (2000).
- American Public Health Association (APHA), American Water Works Association (AWWA) and Water Environment federation (WEF). Standard methods for the examination of water and wastewater (21th ed.). Washington DC: APHA (2005)
- Holt JG, Bergey’s Manual of Determinative Bacteriology, 9th edition (1994).
- Goel PK, Water pollution causes, effects and control. New Age International (P) Ltd., publishers, New Delhi, 269 (1997).
- Shrivastava JN, Verma S, Kumar V, Bioremediation of Yamuna water by mono and dual bacterial isolates. Ind. J. Sci. Res. and Tech. 1(1), 56-60 (2013).
- Prasad MP, Manjunath K, Comparative study on biodegradation of lipid rich wastewater using lipase producing bacterial species. Indian Journal of Biotechnology 10: 121-124 (2011).
- Vasconcellos SP, Cereda MP, Cagnon JR, Foglio MA, Rodrigues RA, Manfio GP, Oliveira VM, In vitro degradation of linamarin by microorganism isolated from cassava wastewater treatment lagoons. Brazilian Journal of Microbiology. 40: 879-883 (2009).
- Ravi Kumar B, Lakshmi Prasad M, Srinivasa Rao D, Sambasiva rao KRS, Bioremediation of sewage using specific consortium of microorganisms. International Journal of Research in Applied, Natural and Social Sciences. 1(6): 15-26 (2013).
- Chin KK, Ong SS, Poh LH, Kway HL, Waste water treatment with bacterial augmentation. J IAEM 22: 50-53 (1995).
- Claxton LD, Houx US, Integration of complex mixture toxicity and microbial analysis for environmental remediation research. Ecotoxicity and Human Health (Lewis Publishers) 467-468 (1995).
- Gaikwad GL, Wate SR, Ramteke DS, Roychoudhury K, Development of Microbial Consortia for the Effective Treatment of Complex Wastewater. J Bioremed Biodeg 5: 227 (2014).
- Zhao S, Hu N, Chen Z, Zhao B, Liang Y, Bioremediation of Reclaimed Wastewater Used as Landscape Water by Using the Denitrifying Bacterium Bacillus cereus. Bull Environ Contam Toxicol 83: 337–340 (2009).
- Mazzucotelli CA, Durruty I, Kotlar CE, Moreira MR, Ponce AG, Roura SI, Development of a Microbial Consortium for Dairy Wastewater Treatment. Biotechnology and Bioprocess Engineering 19: 221-230 (2014).
- Ji-hong Z, Yan L, Zhi-sheng Y, Isolation and characterization of efficient COD degrading bacteria from municipal wastewater. Henan Province Science Foundation for Prominent Youth (2008).
- Rajakumar S,Ayyasamy PM, Shanthi K, Thavamani P, Velmurugan P, Song YC, Lakshmanaperumalsamy P, Nitrate removal efficiency of bacterial consortium (Pseudomonas sp. KW1 and Bacillus sp. YW4) in synthetic nitrate-rich water. Journal of Hazardous Materials. 157:553-563 (2008).
- Krishnaswamy U, Muthuchamy M, Perumalsamy L, Biological removal of phosphate from synthetic wastewater using bacterial consortium. Iranian Journal of Biotechnology 9(1): 37-49 (2011).
- Karthikeyan K, Chandran C, Kulothungan S, Biodegradation of oil sludge of petroleum waste from Automobile service station using selected fungi. Journal of Ecotoxicology and Environmental Monitoring 20: 225-230 (2010).
- Tomar P, Mittal P, Evaluation of Paper Effluent with two Bacterial Strain and their Consortia. International Journal of Education and Science Research Review. 1(2): 25-30 (2014).







