Characterization and Antibiotic Susceptibility Pattern of Bacteria Isolated from Waste Dumping Site in Ujjain City, India
Corresponding author Email: tarunsankhala1@gmail.com
DOI: http://dx.doi.org/10.12944/CWE.16.1.28
Copy the following to cite this article:
Sankhala T, Vyas A, Vyas H. Characterization and Antibiotic Susceptibility Pattern of Bacteria Isolated from Waste Dumping Site in Ujjain City, India. Curr World Environ 2021;16(1). DOI:http://dx.doi.org/10.12944/CWE.16.1.28
Copy the following to cite this URL:
Sankhala T, Vyas A, Vyas H. Characterization and Antibiotic Susceptibility Pattern of Bacteria Isolated from Waste Dumping Site in Ujjain City, India. Curr World Environ 2021;16(1). Available From : https://bit.ly/3l0jqDS
Download article (pdf) Citation Manager Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 08-09-2020 |
|---|---|
| Accepted: | 02-03-2021 |
| Reviewed by: | 
 Rodolfo S Tiburcio
Rodolfo S Tiburcio
|
| Second Review by: |

 Bivek Timalsina
Bivek Timalsina
|
| Final Approval by: | Syed Mohammad Tauseef |
Introduction
This era has witnessed tremendous increase in generation of municipal solid waste. The major reasons for this increase are continuous increase of population, rapid urbanization and escalation in living standards due to the rise of economy1,2. Municipal solid waste (MSW) is a heterogeneous mixture and consists of biodegradable and non biodegradable components. The biodegradable waste mainly comprises food waste, paper, leaves, wood trimming, textile, leather etc., while non biodegradable waste contains plastic, glass, metals etc. Municipal solid waste may also contain hazardous substances like battery, paints, pharmaceutical compounds and e-waste. The composition of municipal solid waste may vary in different areas and depends on living style and socio-economic conditions of the place3. Studies of Gupta and Arora (2016) have indicated that in India 68.8 million tones of municipal solid waste is generated per year. A large portion of municipal solid waste is generated by households which contains biodegradable kitchen waste and non biodegradable materials. The poor management of municipal solid waste results in significant environmental problems and harmful effects on human health5,6. The collection and disposal of Municipal solid waste (MSW) is a major challenge for all developing cities and it is important for protection of the environment and safeguarding human health. In most of the cities the municipal solid waste is collected from different areas of the city and transported to waste dumping grounds. The conventional methods of disposal of this waste include landfill, incineration and composting. The sorting and fractionation of solid waste is an important aspect for management and disposal of this waste. The plastics and non biodegradable materials have to be removed and transported for recycling as they persist in landfill and their incineration is hazardous due to the release of harmful gasses7. A substantial portion of waste remains on ground for spontaneous self degradation. The indigenous bacterial population present at such places plays an important role in spontaneous degradation of the waste. The isolation and characterization of bacteria present at such sites is important for identifying bacteria which can be used for developing bacterial consortium for speeding up the process of waste degradation8. Besides this, many bacteria present at such places can be pathogenic and may be responsible for causing infectious disease and health hazards9. In recent years it has been seen that there is widespread emergence of multidrug resistant bacteria, which is becoming an important constraint in treatment of infectious disease10,11. The objectives of our study were to isolate and identify different bacterial cultures from waste dumping grounds and to determine their antibiotic susceptibility.
Material and Methods
Sample Collection
Soil samples were collected from Kanipura trenching ground and Agar-Maxi bypass trenching ground located near each other on the Agar-Maxi Bypass road in Ujjain city. This site was used for dumping of municipal solid waste of Ujjain city till 2019. Soil samples were collected during different seasons namely, summer (April to June), winter (November to January) and rainy (July to September) over a period of three years (2015 to 2017). In each year three samplings were done and during each sampling six samples from each sampling grounds were collected. During the sampling three samples were taken from the upper layer of soil while three samples were taken from 3 inch below the soil surface. Samples were collected in sterilized zip lock polythene bags with the help of sterile spatula and brought to the laboratory. The samples were stored at room temperature and used for isolation of bacteria within 48 hrs.
Isolation, Purification and Identification of the Bacteria
Bacterial cultures were isolated from soil samples using serial dilution method. One gram of soil was suspended in 100 ml of sterilized distilled water in conical flask. The flask was shaken in order to mix the soil properly. Serial dilutions of this suspension were made (1:10, 1:100, 1:1000, 1:10000 and 1:100000) using sterilized distilled water. Each dilution (0.2 ml) was spread on three different Petri dishes containing Nutrient agar medium using the sterilized glass spreader. The Petri dishes were incubated at 37±2ºC in BOD incubator. Total number of cfu (colony forming units) were counted and it was seen that in the Petri dishes containing dilution 10-1 to 10-3 large number of cfu appeared and the plates were overcrowded, while in Petri dishes containing dilution 10-4/10-5 about 150-225 cfu were obtained. The different looking colonies were isolated and grown in Nutrient agar slants. The bacterial cultures were purified using streak plate method. A small amount of culture was streaked on Petri dishes containing Nutrient agar medium using sterilized wire loop. The Petri dishes were incubated at 37±2ºC in BOD incubator for 48 hrs and well isolated colony was picked and transferred to Nutrient agar slant to obtain pure culture. The cultures were stored in Nutrient agar slants at -20ºC in deep freeze. The cultures were identified on the basis of Gram staining and different biochemical tests including motility, catalase, oxidase, indole, MR, VP, citrate and urease12,13,14.
Antibiotic Susceptibility Pattern
The antibiotic susceptibility pattern of the cultures was determined following Kirby-Bauer disc diffusion antibiotic susceptibility test15. The antibiotics included in the study were Amikacin (30 mcg), Amoxyclave (30 mcg), Ampicillin (10 mcg), Aztreonam (30 mcg), Cefepime (30 mcg), Cefotaxime (30 mcg), Ceftazidime (30 mcg), Chloramphenicol (30 mcg), Ciprofloxacine (5 mcg), Doripenenm (10 mcg), Eratpenem (10 mcg), Gentamicin (10 mcg), Imipenem (10 mcg), Levofloxacin (5 mcg), Piperacillin/ tazobactam (100/10 mcg), Polymyxin B (300 units), Ticarcillin clavulanic acid (30 mcg) and Tobramycin (30 mcg). Distilled water was used as control.
Results and Discussion
In the present study, 63 bacterial cultures were isolated and it was seen that 19.04 percent of the cultures were Gram positive and 80.95 percent of cultures were Gram negative. Different biochemical tests were performed for identification of the cultures and it was seen that 76.19 % of cultures belonged to Pseudomonas sp., 19.05 % of cultures belonged to Bacillus sp. and 4.76 % cultures belonged to Enterobacter sp. This indicates that Pseudomonas is the predominant genus present at this site followed by Bacillus and Enterobacter (Figure 1). These cultures have adapted to grow on these grounds over the years and may be involved in spontaneous degradation of municipal solid waste. Anwar et al., (2017) have proposed that a bacterial consortium of genus Pseudomonas, Bacillus and Seretia may be used to enhance the degradation of kitchen waste. Earlier studies have also shown that Pseudomonas, Bacillus and Enterobacter are important genera isolated from waste dumping grounds; however their frequency varied at different places depending on local conditions16,17,18.
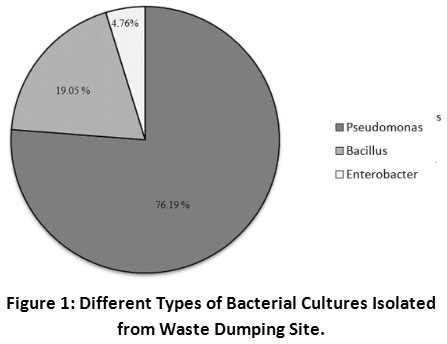 |
Figure 1: Different Types of Bacterial Cultures Isolated from Waste Dumping Site. Click here to view Figure |
The antibiotic susceptibility pattern of 18 antibiotics was determined on all 63 cultures isolated and the percentages of cultures showing resistance to each antibiotic were recorded. Figure 2 shows the percentage of cultures resistant to individual antibiotics. It was seen that 52.38 % cultures were resistant to one or more antibiotics and 47.62 % of cultures were sensitive to all antibiotics tested. Furthermore, 42.85 % cultures were resistant to Aztreonam. The antibiotics Amikacin, Cefepime, Ciprofloxacin and Tobramycin were found to be highly effective as only 4.76 % cultures were resistant to these antibiotics. Ceftazidime, Chloramphenicol, Polymyxin B also showed high effectiveness and 14.28% of cultures demonstrated resistance to these antibiotics. The antibiotic Ticarcillin clavulanic acid showed medium effectiveness and 23.8 % cultures were resistant to this antibiotic. On the other hand, all the cultures were tested to be sensitive to Amoxiclave, Ampicillin, Cefotaxime, Doripenem, Ertapenem, Gentamicin, Imipenem, Levofloxacin and Piperacillin/ tazobactam, indicating that these antibiotics may be used for controlling growth of these bacteria. It was also interesting to note that all the cultures belonging to genera Enterobacter were sensitive to all antibiotics and did not show antibiotic resistance.
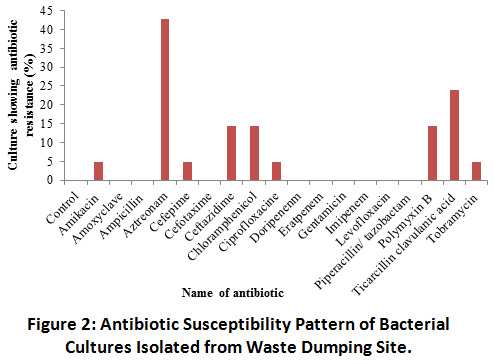 |
Figure 2: Antibiotic Susceptibility Pattern of Bacterial Cultures Isolated from Waste Dumping Site. Click here to view Figure |
The results were also analyzed to determine if the bacteria were resistant to more than one antibiotic. It was seen that 4.76 % of cultures were resistant to 7 antibiotics, 4.76 % of cultures were resistant to 6 antibiotics, 4.76 % of cultures were resistant to 4 antibiotics, 9.52 % of cultures were resistant to 3 antibiotics, 9.52 % of cultures were resistant to 2 antibiotics and 19.04 % of cultures were resistance to single antibiotic. This indicates that about 52 % cultures were antibiotic resistant and about 33 % cultures were resistant to more than one antibiotic. The presence of bacteria showing resistance to more than one antibiotic in significant numbers poses potential risk to public health and may lead to decrease in efficacy of medical treatment10,12. Antibiotic resistant bacteria have been reported from waste dumping ground in various other studies and they have become a global concern10,12,19. It has been suggested that the municipal solid waste landfills and waste dumping grounds provide a suitable environment for emergence of antibiotic resistant bacteria and transfer of resistant genes20. The practice of not sorting municipal solid waste at source leads to indiscriminate disposal of unused/ expired medicines in municipal solid waste and presence of antibiotics and pharmaceutical drugs at such sites creates selective pressure and helps in developing multidrug resistant bacteria due to mutations10,17,21,22. The presence of antibiotic resistant bacteria at such sites may be hazardous to the health of municipality workers, animals and residents of nearby areas. The infections caused by drug resistant bacteria are difficult to treat and increase the cost of medical expenditures17.
Municipal solid waste disposal and its degradation is the basic issue of the current “clean and green” drive of our country. Various applications of microorganisms in management of municipal waste have been examined. Bacterial cultures producing extracellular amylase, cellulase, protease, pectinase and lipase were isolated from waste dumping land of Matigarahat (Siliguri, West Bengal, India) and consortium of these cultures showed good waste degradation ability in laboratory trials8. A compound microbial agent “YH” containing 12 microbial species has been developed in (Beijing, China) having potential application in composing kitchen waste23. Many organizations and researchers throughout the world are working in reducing, recycling and management of this waste, but complete success has not been achieved. Hence, more efforts are needed in this direction so that degradation of municipal solid waste can be accelerated.
Conclusion
In this study, bacterial cultures from waste dumping site of Ujjain have been isolated and identified. It was seen that Pseudomonas is the most frequently occurring bacterial genus present here followed by Bacillus and Enterobacter. It was also revealed that about 52 % bacteria were resistant to antibiotics and about 33 % cultures were resistant to more than one antibiotic. This represents potential risk to the environment and health of animals and humans. Hence, it is suggested that public awareness programs should be conducted, so that waste is sorted in households and pharmaceutical waste is not discarded in municipality waste.
Acknowledgement
We thank Ms. Uzma Choudhary, research scholar in S.S. in Microbiology, Vikram University, Ujjain for her help during the experiments in this work.
Funding Source
The author did not received any funding.
Conflict of Interest
There is no conflict of interest.
References
- Gouveia N, Prado R.R.Health risks in areas close to urban solid waste landfill site, Rev de Saude Publica. 2009;44(5):1-8.
CrossRef - Minghua Z, Xiumin F, Rovetta A, Qichang H, Vicentini F, Bingkai L, Giusti A, Yi L. Municipal solid waste management in Pudong New Area, China, J Waste Manage. 2009;29:1227–1233.
CrossRef - Hussein I, Mona S.M. Solid waste issue: Sources, composition, disposal, recycling, and valorization, Egypt J Pet.2018;27:1275-1290.
CrossRef - Gupta B, Arora S.K. Municipal solid waste management in Delhi-the capital of India, Int J Innov Res Sci Eng Technol. 2016;5(4):5130-5138.
- Palaniveloo K, Amran M.A, Norhashim N.A, Mohamad-Fauzi N, Peng-Hui F, Hui-Wen L, Kai-Lin Y, Jiale L, Razak S.A, Food waste composting and microbial community structure profiling, Processes 2020:8(6), 723; https://doi.org/10.3390/pr8060723.
CrossRef - Ayilara M.S, Olanrewaju O.S, Babalola O.O, Odeyemi O, Waste Management through Composting: Challenges and Potentials, Sustainability 2020, 12, 4456; doi:10.3390/su12114456, 2-23. (Note- These references were old/ incomplete which was pointed by first reviewer so they were revised)
CrossRef - Vucinic A.A, Hublin A, Ruzinski N. Greenhouse gases reduction through waste management in Croatia, Therm Sci. 2010;14(3):681-691.
CrossRef - Sarkar P, Chourasia R.Bioconversion of organic solid wastes into bio fortified compost using a microbial consortium, Int J Recycl Org Waste Agricult. 2017;6:321–334.
CrossRef - Reddy G.R, Pranavi S, Srimoukthika B, Reddy V.V. Isolation and Characterization of Bacteria from Compost for Municipal Solid Waste from Guntur and Vijayawada, J Pharm Sci & Res. 2017;9(9):1490-1497.
- Ifechukwu E, Chioma N, Wesley B, Chituru A, Ijeoma N. Antibiotic resistance pattern of bacterial isolates of liquid wastes and waste dump soils of hospitals in Owerri, Nigeria, Nigerian J Microbiol. 2015;29:3002-3011.
- Ologun O.M, Boboye B, Owoyemi O.O. Molecular Identification and Antibiotic Sensitivity Pattern of Bacteria Associated with Decomposed Domestic Food Wastes from Akure Metropolis, M R J I. 2018;24(3):1-11.
CrossRef - Andy I.E, Okpo E.A. Occurrence and Antibiogram of Bacteria Isolated from Effluent and Waste Dump Site Soil of Selected Hospitals in Calabar Metropolis, Nigeria, M R J I.2018;25(5):1-9.
CrossRef - Begum K, Mannan S.J, Rezwan R, Rahman M.M, Rahman M.S, Nur-E-Kamal A. Isolation and Characterization of Bacteria with Biochemical and Pharmacological Importance from Soil Samples of Dhaka City, Dhaka Univ J Pharm Sci. 2017;16(1):129-136.
CrossRef - Prakasam C, Siddharthan E.P.N, Hemalatha N. Isolation, identification, enumeration and antibiotic profiling of microbes from soil contaminated with hospital waste dumbing, J Pharm Biol Sci. 2017; 5(3):126-133.
- Bauer, A. W., Kirby, Sheries, J. C, Turck, M, Antibiotic. Susceptibility testing by a Standardized single disc method. American J Clini Path. 1996;45:9-58.
- Anwar H, Ashraf M, Saqib M, Ashraf M.A, Ishfaq K. Isolation and screening of biodegrading bacteria from kitchen waste and optimization of physiochemical conditions to enhance degradation, Int J Sci & Eng Res. 2017;8(4):2229-5518.
CrossRef - Borquaye L.S, Ekuadzi E, Darko G, Ahor H.S, Nsiah S.T, Lartey J.S, Mutala A.H, Boamah V.E, Woode E. Occurrence of Antibiotics and Antibiotic-Resistant Bacteria in Landfill Sites in Kumasi, Ghana, J of Chemistry. 2019;1-11.
CrossRef - Mandal C,Tabassum T, Shuvo M.J, Habib A. Biochemical and molecular identification of antibiotic-producing bacteria from waste dumpsite soil, J Adv Biotechnol Exp Ther. 2019; 2(3):120-126.
CrossRef - Sulaimon A.M, Akinwotu O.O, Amoo O.T. Resistance of Bacteria Isolated from Awotan Dumpsite Leachate to Heavy Metals and Selected Antibiotics, Int J Res Pharm and Bioscie. 2015;2(9):8-17.
- Omulo S, Thumbi S.M, Njenga M.K, Call D.R. A review of 40 years of enteric antimicrobial resistance research in Eastern Africa: what can be done better? Antimicrobial Resistance and Infection Control.2015;4(1):02-13.
CrossRef - Efuntoye M.O, Bakare A.A, Sowunmi A.A. Virulence factors and antibiotic resistance in Staphylococcus aureus andClostridium perfringens from landfill leachate, AfricanJ Microbiol Res. 2011; 5(23):3994-3997.
CrossRef - LaPara T.M, Burch T.R, McNamara P.J, Tan D.T, Yan M, Eichmiller J.J. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes in to Duluth-Superior Harbor, Enviro Scie Tech. 2011; 45(22):9543-9549.
CrossRef - Zhao K, Xu R, Zhang Y, Tang H, Zhou C, Cao A, Zhao G, Guo H. Development of a novel compound microbial agent for degradation of kitchen waste, Braz J Microbiol. 2017:48:442-450.
CrossRef







