An Investigation into the Microbiological Quality of Groundwater in an Industrial Hub in Mid-Hills of Northern India
Ajay Kumar Singh1
*
 , Satish Kumar Bhardwaj2
, Satish Kumar Bhardwaj2
 , Rajeev Kumar Aggarwal2
, Rajeev Kumar Aggarwal2
 , Sunita Devi3
, Sunita Devi3
 and Amit Guleri4
and Amit Guleri4

1
Department of Health and Family Welfare,
Solan,
Himachal Pradesh
India
2
Department of Environmental Science,
College of Forestry, YS Parmar University of Horticulture and Forestry, Nauni,
Solan,
Himachal Pradesh
India
3
Department of Basic Sciences,
YS Parmar University of Horticulture and Forestry, Nauni,
Solan,
Himachal Pradesh
India
4
Department of Economics and Sociology. Economist (QM),
College of Basic Sciences and Humanities, Punjab Agricultural University,
Ludhiana,
Punjab
India
Corresponding author Email: ajaysingh7279@gmail.com
DOI: http://dx.doi.org/10.12944/CWE.18.3.16
Copy the following to cite this article:
Singh A. K, Bhardwaj S. K, Aggarwal R. K, Devi S, Guleri A. An Investigation into the Microbiological Quality of Groundwater in an Industrial Hub in Mid-Hills of Northern India. Curr World Environ 2023;18(3). DOI:http://dx.doi.org/10.12944/CWE.18.3.16
Copy the following to cite this URL:
Singh A. K, Bhardwaj S. K, Aggarwal R. K, Devi S, Guleri A. An Investigation into the Microbiological Quality of Groundwater in an Industrial Hub in Mid-Hills of Northern India. Curr World Environ 2023;18(3).
Download article (pdf) Citation Manager Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2023-05-04 |
|---|---|
| Accepted: | 2023-08-21 |
| Reviewed by: | 
 Pramod G
Pramod G
|
| Second Review by: |

 Maheswari Sundararaman
Maheswari Sundararaman
|
| Final Approval by: | Dr. Gopal Krishan |
Introduction
Globally, various anthropogenic activities related with industrialization, urbanization and chemical based farming have affected the physical, chemical and biological properties of drinking water1, 2. Polluted water transmits various infectious diseases such as shigelosis, dysentery, diarrhoea, cholera, hepatitis etc3, 4. Poor water sanitation and hygiene is responsible for about 88% of the diarrhoeal disease across the world5.
In India, anthropogenic activities lead to about two million tons of human waste into water courses daily6. District Solan, a rapidly industrializing region in Himachal Pradesh, a state in northern India, has also witnessed rapid growth in urbanization and industrialization which has led to unabated pollution of water7, necessitating water quality assessment.The utility of the thermotolerant coliform group, specifically Escherichia coli, as an indicator of microbiological water quality, is definitive evidence of faecal contamination8.
Sometimes, the sole identification of indicator organism can be misleading. Therefore, many advanced microbiological analytical (DNA-based typing methods) tests have been used recently9. The use of rRNA molecules was developed for comparisons based on phylogenetic traits10. The16S rRNA gene (1550 bp) has been the most used marker11. Henceforth, the study was undertaken with the objective of the microbiological assessment of region’s drinking water sources by looking for the presence of indicator organisms, the thermotolerant coliforms and further identification of the organism by genome typing.
Materials and Methods
In order to construct the water borne disease burden strata of the district, the standard stratification method was employed to the five-year diarrhoeal disease incidence secondary data12. This was collected from the district health department for the 49 PHIs (Public Health Institutes) of the district for the years 2012-1613. Approximation statistics were applied and disease burdened areas were divided in four strata namely Very low, Low, High, and Very high. The Public Health Institute (PHI) Nalagarh of industrial hub located in Shivalik foothills of North Western Himalayas of district Solan, India, having high disease burden, was selected for assessing the microbiological attributes of the sources of drinking water.
Hydrogeology of study region
In the study region, groundwater occurs in unconsolidated alluvial formation comprising sand, silt, gravel, cobbles/pebbles. In Nalagarh, wells and tube wells are the main groundwater obstruction structures. Nalagarh has dendrite drainage pattern having sandstaone-clay alteration. The uppermost horizon consists of conglomerate with well-rounded clast of grey quartizite possibly derived from shale.
Study design and data sampling for microbiological assessment
A cross-sectional survey was conducted in the industrial region being served by PHI Nalagarh. KAP attributes (Knowledge, Attitude and Practice) of 180 adult residents was assessed for water, sanitation and hygiene. A structured pre-tested standardized questionnaire was used. Five commonly used sources of water were inferred from survey. These sources (three bore-wells and two unprotected springs) were selected purposively for the further study (Fig.1). The groundwater samples were drawn by the grab water sampling technique. The sampling was undertaken during the monsoon months of June, July and August and during the post monsoon months of September, October and November in 2018-1914.
Detection of thermotolerant coliforms
For determining the presence of thermotolerant coliforms, the technique of multiple tube fermentation was used 14. Appropriate decimal dilutions of water sample were inoculated in a series of tubes. Three stage procedure was employed for identifying aerobic and facultative anaerobic rod shaped bacteria which were rod shaped, non spore forming and gram negative. These bacteria (coliforms) fermented lactose with acid and gas formation within 48 hours at 35 ± 2oC,. Most Probable Number (MPN) per 100ml water was used to represent the results.
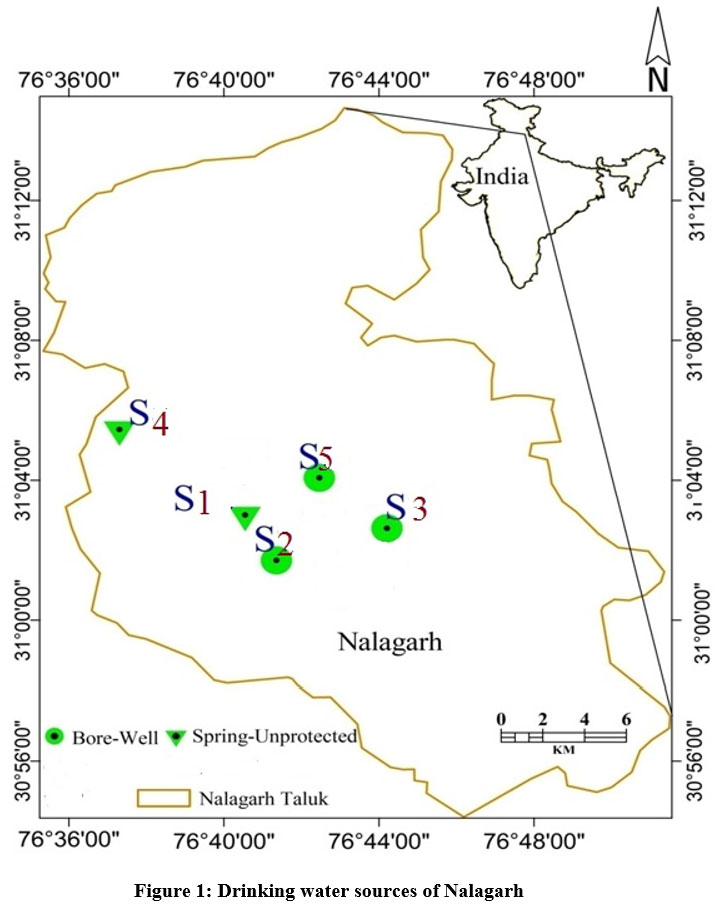 | Figure 1: Drinking water sources of Nalagarh
|
Morphological and biochemical characterization
Different morphotypes of coliform bacteria obtained on Eosin Methylene Blue (EMB) agar plates in the present study were identified based on their morphological and biochemical characteristics according to the standard methods described in Bergey, Manual of Systematic Bacteriology15.
Molecular characterization
Molecular characterization of the most prevalent indicator organism was done using 16S rRNA gene sequencing as per the standard protocol (Fig.2). The isolate was identified at National Centre for Cell Science Pune, Maharashtra, India. Genomic DNA was isolated by phenol/chloroform extraction method. Thereafter, 16S rRNA gene amplification by PCR was achieved by employing the universal primers such as 16F27 [5'-CCA GAG TTT GAT CMT GGC TCA G-3'] and 16R1492 [5'-TAC GGY TAC CTT GTT ACG ACT T-3']. For purifying the amplified 16S rRNA gene PCR product the method of PEG-NaCl precipitation was adopted. Thereafter, it was directly sequenced on an ABI® 3730XL automated DNA sequencer. For sequencing, additional internal primers were utilized from both the ends ensuring that each position was read at least twice. Thereafter, Lasergene package was used for assembly. EzBioCloud database (http://ezbiocloud.net/eztaxon) was then employed for identification. MEGA 7 software programme was used for phylogenetic analyses. The gene sequences of 16S rRNA were deposited in the Gene Bank ofNCBI(National Centre for Biotechnology Information), USA.
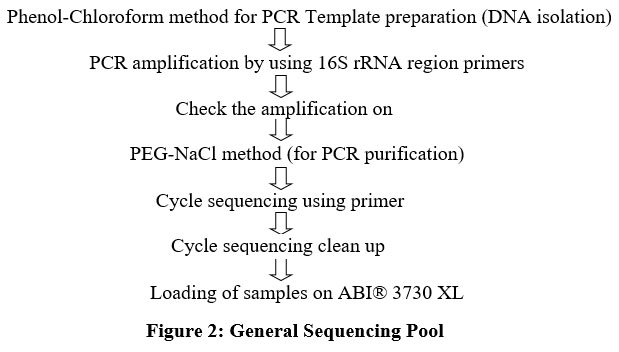 | Figure 2: General Sequencing Pool
|
Deposition of bacterial strain NGW
The bacterial strain Raoultella planticola NGW, identified in the present study was deposited in a national culture collection centre/repository namely the National Centre for Microbial Resource and the National Centre for Cell Science at Pune in Maharashtra state of India, for making its accessibility easy as reference strain by the research community.
Analysis
Randomized Block Design (Factorial ) suggested by Gomez and Gomez was used for data analysis16. Five water sources were presumed to be the replications for the study. The years were considered as factor one with two levels i.e. 2018 and 2019. Seasons were factor two having monsoon and post-monsoon as two levels. Least Significant Difference (LSD) at 5 % level of probability was utilized for comparing the treatment means.
Results and Discussion
Table 1 inferred MPN values of thermotolerant coliforms. It varied from 2-34 per 100 ml water across all the water sources in both seasons. This indicated varying contamination levels in drinking water of the region. the seasons (monsoon and post-monsoon) significantly influenced the concentration of thermotolerant coliforms in water. These bacteria were detected more in concentrations during the monsoon season (p = 0.02, 95% confidence interval). However, the concentration of the coliforms was not influenced by the year of the study. Also, no effect on the concentration of the coliforms occurred due to any interaction between the two factors i.e. the year and the season. The average highest total coliforms (MPN- 27 per 100 mlwater)were detected in the water sources during monsoon, 2019. This was followed by statistically equivalent values noticed during the monsoon, 2018.
Morphological characterization of coiliform morphotypes
A total of four different morphotypes of coliform bacteria were obtained from the five different drinking water sources of industial region Nalagarh on EMB agar plates. All morphotypes were further purified on EMB agar plates. Of these, only one isolate, designated as NGW (Nalagarh Water) strain showed green metallic sheen on EMB plates (Fig. 3 and Table 2). During the monsoon season, NGW strain was detected in all the water sources. This suggested the prevalence/dominance of NGW strain among other strains, in the selected water sources. On the contrary, NGW strainwas observed in only two water sources during post monsoon
Table 1: Temporo-spatial distribution of thermotolerant coliforms in drinking water sources of Nalagarh.
Year | Season | Water sourcea (MPN per 100 ml water) | Mean | Yearly mean | ||||
A | B | C | D | E | ||||
2018 | Mb | 17 | 34 | 33 | 33 | 17 | 26.80 | 19.00 |
PMc | 17 | 17 | 9 | 9 | 4 | 11.20 | ||
2019 | M | 17 | 34 | 34 | 33 | 17 | 27.00 | 18.50 |
PM | 12 | 17 | 2 | 2 | 17 | 10.00 | ||
Least Significant Difference (0.05) Year: Not significant; Season: 7.48; Year x Season: Not significant | ||||||||
aDrinking water sources of : A-Ward 1; B-Ward 3; C- Near bus stand; D- Ward 7; E- Raj Mahal area, bMonsoon, cPost monsoon
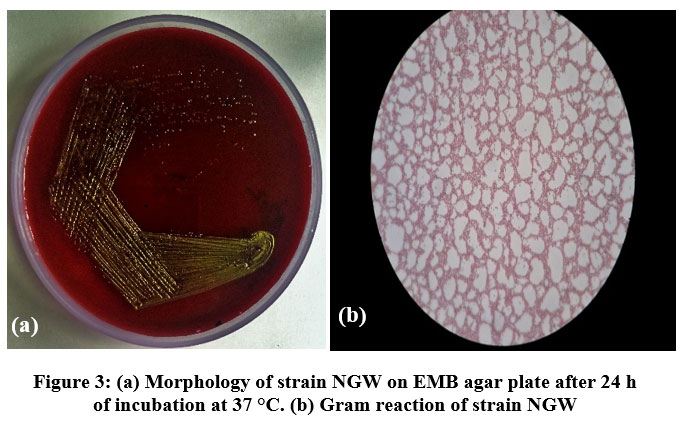 | Figure 3: (a). Morphology of strain NGW on EMB agar plate after 24 h of incubation at 37 °C. (b). Gram reaction of strain NGW
|
Table 2: Phenotypic characterization of Nalagarh water strain (NGW) strain
| Phenotypic characterization | ||||||
| Morphological characteristics (on Eosin Methylene Blue agar plate) | ||||||
Colour
| Margin | Shape | Texture | Elevation | Gram reaction | Metallic sheen- |
Greenish | Entire | Circular | Smooth | Flat | Negative | Green |
| Biochemical characteristics | ||||||
Positive reaction for - Methyl Red, oxidase, catalase, oxidative, triple iron sugar tests | Negative reaction for - Voges- Proskauer, indole, citrate and gelatinase tests | Positive reaction for carbohydrate fermentation (Acid/Gas production) -Arabinose, Mannose, Mannitol, Xylose, Rhamnose, Dextrose, Sorbitol, Lactose, Adonitol, Melibiose, Cellobiose and Sucrose | Probable identification- Escherichia coli | |||
Table 3: Molecular characterization of Nalagarh water strain (NGW) strain.
Molecular characterization | |||
NCBI GenBank Accession number
| Closest neighbour | Percent Similarity
| Sequence length (bp)
|
MK318824 | RaoultellaplanticolaATCC 33531 (T) | 99.85 | 1200 |
Biochemical characterization
Based on biochemical characteristics, strain NGW was positive for MR, catalase, oxidase, O/F and TSI tests whereas, it showed negative reactions for VP, Indole and citrate tests (Fig. 4 and Table 2). All morphological characterization results, especially green metallic sheen formation on EMB agar plates and biochemical tests except indole tests, prompted the possibly identification of this strain as Escherichia coli.
.jpg) | Figure 4: Biochemical characteristics of NGW strain.
|
Molecular characterization
16S rRNA ribotyping of the NGW strain revealed that it belonged to the genus Raoultella Your sentence may be unclear or hard to follow. Consider rephrasing. and species planticola, GenBank accession no: MK318824 (Table 3). The sequence comparison with 16S rRNA gene sequences deposited in the GenBank indicated that nearest neighbors of strain NGW were Raoultella planticola ATCC 33531 (T), Raoultella ornithinolytica JCM 6096 (T), Raoultella electrica1GB (T) (AB762091), Enterobacter aerogenes KCTC 2190 (T) (CP002824) and Klebsiella grimontii 06D021 (T) (FZTC01000044). Phylogenetic tree constructed with Neighbor-Joining method interpreted that the strain NGW clustered among species of genus Raoultella. Henceforth, it was confirmed by the observations that the strain NGW belonged to the genus Raoultella (Fig. 5).
It was observed in the present study that the concentrations of thermotolerant coliforms in the drinking water sources was high. Similar findings had been also ascertained by a KAP study undertaken in these regions by Singh and Bhardwaj2. Bain et al. had demonstrated that the chance of faecal contamination were more in the unprotected water bodies and that the odds of contamination in protected sources were lower (OR = 0.15) when compared17 Nalagarh region had predominantly bore wells and unprotected springs as the source of drinking water. The present study has inferred high contamination levels in these sources. In Baddi region, which was adjoining the study area, another study had documented high coliform concentrations in bore-well water18. Poor water quality due to adverse bacteriological profile has been observed in water samples of Shimla, the town in the vicinity of our study area19, 20. The present study documented that drinking water sources were unprotected and prone to runoff and faecal contamination. A study in West coast of India had similarly documented high densities of coliforms in monsoon seasons21. Another similar study had inferred that the concentration of total coliforms and E. coli was more in the wet season as compared to the dry season 22. The water bodies of industrially dominated regions had shown E. coli, suggesting lack of sanitation services. This was leading to faecal contamination of these water sources of the region. The migratory populations of various industries practiced open defecation. This feature had also been observed during the KAP survey carried out in the present study. A similar finding wass observed in a study conducted Taiwan which demonstrated that the domestic sewage of areas having a large number of industries, was responsible for high concentrations of faecal coliforms23. The study elicited low prevalence of Raoultella planticola in post monsoon as compared to monsoon season. In another similar study, conducted in the northeast state of Odisha, India, seasonal variation of E.coli fecal indicator organism was documented and low prevalence was observed in post monsoon season24.
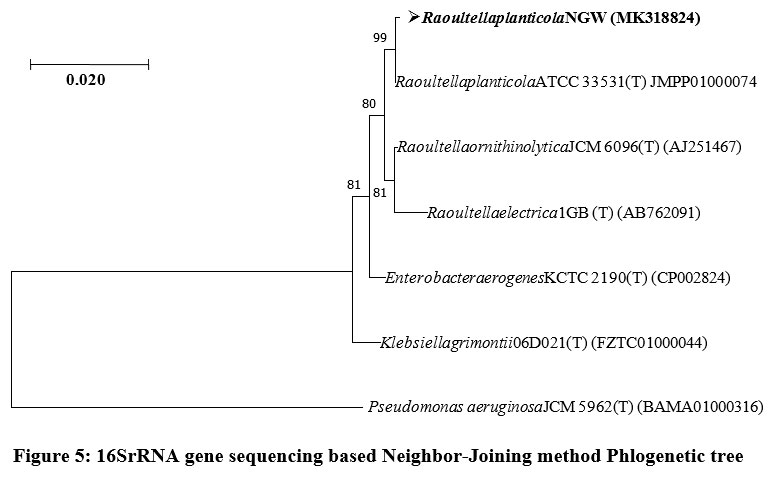 | Figure 5: 16SrRNA gene sequencing based Neighbor-Joining method Phlogenetic tree.
|
It was demonstrated in the study that the thermotolerant coliforms are the indicator organisms which can be utilized for the sanitary quality assessment of drinking water sources. The presence of thermotolerant coliforms pointed out to the probable fecal contamination of water. Similarly, Pandey and Soupir (2013) had also demonstrated water having varying levels of E. coli and studied utility of E. coli for water quality management25.
The present study has concluded that the indentification of indicator organism i.e. E. coli, based upon phenotypic identification methods is not sufficient. An exact indentification of the micorganism involved in the contaminated water may be inferred only after detailed genotypic tests, as was the strain Raoultella planticola identified in our study. Primarily R. planticola is found in water and soil. This rod shaped bacteria is gram-negative, aerobic, encapsulated and nonmotile in nature. The bacteria is a causative agent for urinary tract infections, cellulitis, necrotizing fasciitis, bacteremia, soft tissue infections, conjunctivitis, peritonitis and pneumonia. It also causes conjunctivitis, pneumonia, cholangitis, urinary tract infection, peritonitis, cellulitis, and soft tissue infection26. R. planticola was initially included in the genus Klebsiella. In 1981, it was first described as Klebsiella planticola 27and as Klebsiella trevisanii in 198328. Comparative analysis of 16S ribosomal RNA and rpoB gene sequences identified Raoultella which according to current taxonomy, has been described as a new genus of the family Enterobacteriaceae 28.
Furthermore, phenotypic identification results revealed that the NGW strain may belong to E.coli while genotypic identification confirmed its identification as Raoultella planticola (formerly Klebsiella planticola). The strain identified has been documented elsewhere in recent studies as a new pathogen afflicting human health30.This discrepancy in results could be ascribed to the fact that identification systems based upon biochemical methods may sometimes be unreliable for environmental isolates as the computerized databases do not have sufficient information about these environmental bacteria. Similarly a study on the eggs of loggerhead sea turtle, biochemical (API and Microgen) and molecular methods (16S rRNA analysis) employed in the identification of bacterial isolates inferred a 74 percent discrepancy in the identification results31.
This type of discrepancy was also observed in a study which reported that physiological tests have certain uncertainity in the identification of E. coli32. Escherichia coli has been confirmed by the characteristics such as gas and indole formation at 44 and 44.5 ?C respectively. This feature also makes it distinct from Klebsiella species. Similarly another study underlined the use of alternative microbial techniques for real identification of contaminant microorganism33.
The study indicated that the presence of thermotolerant organisms preliminarily evinced contaminated water sources. Genotypic molecular studies elucidated that the water sources used for drinking purposes, of the industrial region of district Solan, were contaminated by the organism Raoultella planticola. Henceforth, the bore well and the unprotected spring water sources need to be managed to check the contamination by this rare human pathogen. The town administration was appraised of the findings of the study and remedial actions thereafter had been put in place for the treatment of water sources.
Acknowledgement
The author would like to thank Department Environmental Science, YSPUHF Nauni, for providing logistic support for the study.
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Funding Sources
There was no financial support/funding for this research work.
References
- Matache M. L, David I. G, Matache M, Ropota M. Seasonal variation in trace metals concentrations in the Lalomita river, Romania. Environ Monit Assess 2009; 153:273-79. https://doi.org/10.1007/s10661-008-0354-y
CrossRef - Singh A. K, Bhardwaj S. K. Assessment of spatial and temporal variation of water quality in mid hills of North Western Himalayas- a water quality index approach. Curr World Environ2019; 14:37-48. https://doi.org/10.12944/CWE.14.1.06
CrossRef - Giwa S. I, Abubakar M. G, Abdullahi Z, Ishaq D. U. Determination of lead concentration in different water sources collected from Sokoto metropolis, Nigeria. IOSR- JAC2014; 7:53-56.
- Smith R. S. Classification of water related disease.Water and Health. In: Encyclopedia of Life Support Systems; 2018: 21pp. https://www.esolss.net/Sample-Chapters/C03/E2-20A-01-01.pdf
- WHO. Time For Global Action For People And Planet. United Nations The Millennium Development Task Force Report. New York; 2015:7pp. https://www.un.org/millenniumgoals/2015_MDG_Report/pdf/MDG%202015%.pdf. Accessed 23 July 2020
- Mensah F. O, Alo C, Yidana S. M. Evaluation of ground water recharge estimates in a partially metamorphosed sedimentary basin in a tropical environment: application of natural tracers. Sci. World J. 2014; 8pp,Article ID:419708. https://doi.org/10.1155/2014/419508
CrossRef - Singh A. K, Bhardwaj S. K, Devi S. Microbiological status of drinking water sources and its relationship with human health in Solan, India. Environ Monit Assess 2021; 193:32. https://doi.org/10.1007/s10661-020-08833-x
CrossRef - Thomas J. C, Lutz M. A, Bruce J. L, Graczyk D. J, Richards K. D, Krabbenhoft D. P et al. Water quality characteristics for selected sites within the Milwaukee Metropolitan Sewerage District Planning Area, Wisconsin, February 2004-September 2005. Scientific Investigations Report; 2007. https://pubs.usgs.gov/sir/2007/5084page9.html
CrossRef - Price R. G, Wildeboer D. E. coli as an indicator of contamination and health risk in environmental waters. Escherichia coli- recent advances on physiology, pathogenesis and bacteriological applications. Amidou Samie, IntechOpen2017; https://doi.org/10.5772/67330
CrossRef - Woese C. R. Bacterial evolution. Microbiol. Rev 1987; 51:221-271. https://www.pubmed.ncbi.nlm.nih.gov/2439888
CrossRef - Amann R. I, Ludwig W, Schleifer K. H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev1995; 59:143-169. https://www.pubmed.ncbi.nlm.nih.gov/7535888
CrossRef - Singh R, Sukhatme B. V. Optimum Stratification. Ann Inst Stat Math 1969; 21:515-528. https://doi.org/10.1007/BF02532275
CrossRef - Anonymous. 2016. Integrated Disease Surveillance Programme. Disease alerts/ outbreaks reported and responded to the state/UTs through integrated disease surveillance project. Available at www.idsp.nic.in
- APHA. Standard methods for the examination of water and wastewater (24thed).Washington D.C; 2017: 1496 pp.
- Vos P, Garrity G, Jones D, Krieg N. R, Ludwig W, Rainey F. A et al. The Firmicutes Volume 3. Bergey’s Manual of Systematic Bacteriology,Williams and Wilkins, 1984; 2009. https://www.springer.com/gp/book
- Gomez K. A, Gomez A. A. Statistical procedures for agricultural research with emphasis on rice. John Wiley and Sons, New York; 1984: 680pp. Accessed 23 July 2020
- Bain R, Cronk R, Wright J, Yang H, Slaymakr T, Bartram J. Fecal contamination of drinking- water in low- and middle- income countries: a systematic review and meta-analysis. PLoS Med2014; 11: e1001644. https://doi.org/10.1371/journalmed.100164
CrossRef - Karnwal A, Dohro A, Mannan M. A. Microbial analysis of potable water and its management through useful plant extracts. Int.j.sci.res2017; 73:44-49. https://doi.org/10.21506/j.ponte.2017.3.34
CrossRef - Bhagra S, Singh D, Sood A, Kanga A. Bacteriological profile of water samples in and around Shimla hills: a study from the sub Himalayan region. Int J Community Med Public Health2017; 4:1966-1971. https://doi.org/10.18203/2394-6040.ijcmph20172158
CrossRef - Ganju S. A, Gautam N, Walia S, Kanga A. Seroepidemiology of a recent outbreak of Hepatitis E in urban Shimla, Himachal Pradesh, India. J Commun Dis 2017; 49:17-22. https://doi.org/10.24321/0019.5138.201709
CrossRef - Borade S, Dhawde R, Maloo A, Syed N, Dastager G. Occurrence and seasonal variation in distribution of fecal indicator bacteria in Tapi estuary along the West coast of India. Indian J Mar Sci2014; 43:340-347. https://nopr.niscair.res.in/handle/123456789/27338
- Hong H, Qiu J, Liang Y. Environmental factors influencing the distribution of total and fecal coliform bacteria in six water storage reserviours in the Pearl river delta region, China. J. Environ. Sci (China)2010; 22:663-668. https://doi.org/10.1016/s1001-0742(09)60160-1
CrossRef - Liou S. M, LO S. L, Wang S. H. A generalized water quality index for Taiwan. Environ Monit Assess.2003; 96:35-52.
CrossRef - Banoo S, Kar R. N, Panda C. R. Seasonal variation and distribution of sewage pollution indicator and human pathogenic bacteria along Odisha coast. Indian J Mar Sci 2013; 43: 859-869. https://nopr.niscair.res.in/handle/123456789/28771
- Pandey P. K, Soupir M. L. Assessing the impacts of E. coli laden streambed sediment on E. coli load over a range of flows and sediment characteristics. J Am Water Resour Assoc 2013; 43: 1261-1269. https://doi.org/10.1111/jawr.12079
CrossRef - Mehmood H, Pervin N, Haq MI, Kamal KR, Marwat A, Khan M. A rare case of Raoultella planticola urinary tract infection in a patient with immunoglobulin A nephropathy. J. Investig. Med. High Impact Case Rep 2018; 6: 2324709618780422. https://doi.org/10.1177/2324709618780422
CrossRef - Bagley S. T, Seidler R. J, Brenner D. J. Klebsiella planticola SP nov.: a new species of Enterobacteriaceae found primarily in nonclinical environments. Curr. Microbiol 1981; 6:105-109. https://doi.org/10.1007/BF01569013
CrossRef - Ferragut C, Izard D, Gavini F, Kersters K, Lay J. D, Leclerc H. Klebsiella trevisanii: a new species from water and soil. Int. J. Syst. Evol. Microbiol 1983; 33: 133-142.
CrossRef - Drancourt M, Bollet C, Carta A, Rousselier P. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb.nov.Int. J. Syst. Evol 2001; 51: 925-932. https://doi.org/10.1099/00207713-51-3-925
CrossRef - Atrici S, Unkar Z. A, Demir S. O, Akkoc G, Yakut N, Yilmaz S, et al. A rare and emerging pathogen: Raoultella planticola identification based on 16S rRNA in an infant. J. Infect. Public Health2017; 11:13-132. https://doi.org/10.1016/j.jiph.2017.03
CrossRef - Awong-Taylor J, Craven K. S, Griffiths L, Bass C, Muscarella L. Comparison of biochemical and molecular methods for the identification of bacterial isolates associated with failed loggerhead sea turtle eggs. J. Appl Microbiol 2007; 104:1244-1251. https://doi.org/10.1111/j.1365-2672.2007.03650.x
CrossRef - Niemi R. M, Mentu J, Siitonen A, Niemela S. I. Confirmation of Escherichia coli and its distinction from Klebsiella species by gas and indole formation at 44 and 44.5 C. J. Appl. Microbiol 2003; 95:1242-1249. https://doi.org/10.1046/j.1365-2672.2003.02125.x
CrossRef - Schriewer A, Odagiri M, Wuertz S, Misra P. R, Panigrahi P, Clasen T, Jenkins M. W. Human and animal fecal contamination of community water sources, stored drinking water and hands in rural India measured with validated microbial source tracking assays. Am. J. Trop. Med. 2015; 93:509-516. https://doi.org/10.4269/ajtmh.14-0824
CrossRef







