Biodegradation of formaldehyde under saline conditions by a moderately halophilic bacterial consortium
Krishnaswamy Veenagayathri1 * and Namasivayam Vasudevan1
DOI: http://dx.doi.org/10.12944/CWE.5.1.05
A bacterial consortium isolated from saline environment was used for in the degradation of formaldehyde under saline conditions. Formaldehyde degradation was studied with the bacterial consortium at various NaCl concentrations from 3 % to 7%. The consortium utilized formaldehyde (100-400mg/L), and degraded within 96 h. The degradation of formaldehyde was optimum at 5 % NaCl (w/v). The removal efficiency of formaldehyde could be enhanced by the addition of growth promoting substances namely yeast extract, tryptone and urea, of which yeast extract proved to be efficient. The presence of different salts (i.e. KCl, Na2SO4, K2SO4 and NaNO3) did not affect the microbial growth and biodegradation of formaldehyde. The consortium could degrade formaldehyde along with phenol as the co-substrate.
Copy the following to cite this article:
Veenagayathri K, Vasudevan N. Biodegradation of formaldehyde under saline conditions by a moderately halophilic bacterial consortium. Curr World Environ 2010;5 (1):31-38 DOI:http://dx.doi.org/10.12944/CWE.5.1.05
Copy the following to cite this URL:
Veenagayathri K, Vasudevan N. Biodegradation of formaldehyde under saline conditions by a moderately halophilic bacterial consortium. Curr World Environ 2010;5 (1):31-38. Available from: http://www.cwejournal.org/?p=1094
Download article (pdf)
Citation Manager
Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2010-04-25 |
|---|---|
| Accepted: | 2010-05-20 |
Introduction
Formaldehyde is an important intermediate compound used in a large variety of processes in the chemical industry. It is frequently found in wastewaters and waste gases causing environmental pollution Prado et al., (2003). Formaldehyde is also used in the industrial processing of wood, paper production, leather, resin and glue (Krick et al., (1980)). Formaldehyde is found in wastewaters in monomeric (free) form, as well as its derivatives such as ether, urea, phenolic condensates and also forms composite complexes with resins, phenols and many other chemicals. Most of the formaldehyde derivatives are generated in chemical industrial processes as waste or by-products and they are much less susceptible to biodegradation than free formaldehyde.
Formaldehyde is often used as an active ingredient in preservatives and disinfectants to inhibit microbial activity because of its biocidal property. It is a highly reactive toxic compound which can cause severe action on living organisms. Despite its microbiocidal activity, formaldehyde can be assimilated by methylotrophic microorganisms as a sole source of carbon and energy and it is a key metabolic intermediate in the oxidation of methanol. Bonastre et al., (1986) reported the partial biodegradation of formaldehyde at a concentration of 2300 mg/L with activated sludge. Adroer et al., (1990) reported that Pseudomonas putida A2 degraded 400 mg/L of formaldehyde as the sole carbon source in batch study.
Few studies have also been conducted on the degradation of formaldehyde by Pseudomonas (Kato et al., (1983), Yanase et al., 1995) Escherichia coli (Gutheil et al., (1997), various strains of Methylotropha Attwood and Quayle (1984), Hansenula sp. Kaszycki, and Koloczek (2000) and Trichosporum sp. (Kaneko 1985). Azachi et al., (1995) found that Halomonas sp. MA-C was able to degrade 75-100 mg/L formaldehyde in a salt medium and the isolate transformed formaldehyde at a high rate by means of a constitutive NAD- and glutathione-dependent formaldehyde dehydrogenase. Yamazaki et al., (2001) studied the biodegradation of formaldehyde by a formaldehyde-resistant bacterium DM-2 strain which was isolated from coastal seawater. DM-2 strain pre-cultured in the presence of formaldehyde was able to completely degrade 400 mg/L formaldehyde in the mineral medium in the presence of 3 % NaCl. Hidalgo et al., (2002) reported formaldehyde removal in a synthetic medium and industrial wastewater by Rhodococcus erythropolis UPV-1.
In saline wastewaters, formaldehyde is discharged from different industries like phenol-resin manufacturing industry (Cvetkovic et al 1988), and urea-formaldehyde adhesives industry (Vidal et al., 1999). Formaldehyde biodegradation was attempted in non- halophilic conditions, whereas biodegradation of the substrate has not been reported under halophilic conditions. Here we have report the isolation of a halotolerant bacterial consortium from saline environment for the degradation of formaldehyde.
Materials and Methods
Bacterial Consortium and Culture Conditions
The bacterial consortium was isolated from soil samples from different habitats of Chennai, having proximity to saline environment. During the initial adaptation stage the consortium was enriched with phenolic compounds and they were biochemically characterized, having 6 strains, of which 4 strains were gram positive and two strains are gram negative. Further analyses with 16S rRNA gene sequence analysis, 6 isolates were identified as Bacillus cereus, Arthrobacter sp., Bacillus licheniformis, Halomonas salina, Bacillus subtilis and Pseudomonas aeruginosa. The sequences of the isolates were deposited in GenBank and their accession numbers are EU780459, EU780460, EU780461, EU780462, EU780463, and EU780464.
During the enrichment period with formaldehyde, the consortium was acclimatized with 100mg/L of formaldehyde as the sole carbon and energy source. The bacterial consortium was enriched in mineral salts medium (g/L) of NaCl, 30.0 or 50.0 or 70.0, KH2PO4 0.25, NH4Cl 1.0, Na2BO7 2.0, FeCl3 0.0125, CaCl2 0.06 and MgCl2 0.05. The medium was supplemented with a specified amount of added NaCl and 10 mg of yeast extract, adjusted to pH -7 (Alva and Peyton 2003). The medium was autoclaved, cooled to room temperature and was amended with formaldehyde (100 mg/L) through a sterile filter (0.45 µm) in 250ml Erlenmeyer flasks. The chemicals and reagents used in the study were analytical grade.
Growth and Degradation of Formaldehyde
Growth of the bacterial consortium on formaldehyde was studied by determining the colony forming units (CFUs) per ml on nutrient agar medium. The mineral medium was substituted with different concentrations of NaCl (1%, 3 %, 5% and 7%) and formaldehyde from 100-400 mg/L. Initially the medium was supplemented with 100 mg/L of the substrate, from this 5ml aliquots was aseptically inoculated into 100ml of liquid medium supplemented with increasing formaldehyde concentrations. At 24h time interval, the culture was centrifuged at 10,000 rpm for 15 minutes to remove the cells. The culture supernatant was collected and used for the estimation of residual formaldehyde.
Biodegradation on Formaldehyde
For the degradation study, mineral salts medium containing formaldehyde was inoculated with the bacterial consortium Different conditions used for the degradation of formaldehyde were (i) medium + formaldehyde + bacterial consortium (ii) medium + formaldehyde and (iii) medium + bacterial consortium, with (ii) and (iii) serving as controls. The bacterial consortium was added to the medium at concentrations of 104– 105 cfu/mL. The culture, in duplicate, was incubated at 37 °C with shaking at 150 rpm and extracted every 24 h interval for 5 days.
Determination of Formaldehyde
Formaldehyde determination was performed by the Hatzsch method (Nash 1953, Doromina et al., 1997). Five milliliter of the culture sample was mixed with equal volumes of Hantzch reagent (2 M ammonium acetate, 50mM acetic acid, 20mM acetyl acetone) and incubated at 60 °C for 10 minutes. The resulting yellow color was measured at 412 nm
Gas Chromotographic Analysis for Phenol Estimation
To estimate the amount of phenol, gas chromatographic analysis was used. The cell suspensions were clarified by centrifugation at (10,000 rpm for 15 min, 6°C). The culture supernatant was extracted with dichloromethane, condensed and filtered through a 0.2mm Gelman filter acro disc, prior to analysis in gas chromatograph (Chemito GC Model No 1000) equipped with FID detector and capillary column (Varian Chromopak capillary column CP SIL 8 CB, 30m X 0.32 mm . Nitrogen was used as a carrier gas, injector temperature was 220°C, detector temperature was 250°C and the oven temperature of the column was maintained at 150°C.
Effect of pH, Salts and Nitrogen Source
The effect of different pH (6, 7 and 8) was examined with the bacterial consortium with 100mg/ L formaldehyde at 5 % NaCl. The growth medium was supplemented with different nitrogen sources such as yeast extract (0.01 %), tryptone (0.01 %) and urea (0.01 %). Effects of alternate salts on the biodegradation of formaldehyde were determined by addition of the 5 % individual salts such as KCl, Na2SO4, K2SO4 and NaNO3
Effect of Co-Substrate
To study the effect of co-substrate, 100 mg/L of phenol was used as the co-substrate along with formaldehyde (100 mg/L) at 5 % NaCl and the degradation efficiency was studied.
Results and Discussion
Formaldehyde Degradation of Various NaCl Concentrations
The consortium containing six strains were enriched from soils from saline environment was used for degradation of formaldehyde. The ability of the consortium to degrade formaldehyde (100 mg/L) at various salt concentrations (1%, 3%, 5%, and 7% NaCl) was investigated. Salinity of the medium influenced the degradation of formaldehyde and growth of the consortium was optimum at 5 % NaCl. When the salinity of the mineral medium was increased to 7 % the removal efficiency of formaldehyde reduced. The growth of the consortium increased from 4 x 103 to 7 x 107 in case of 5 % NaCl, complete degradation of formaldehyde was achieved within 96 h. Growth of the bacterial consortium and degradation efficiency of formaldehyde (100 mg/L) at different NaCl concentrations is presented in (Figure1).
Earlier studies have used individual strains for the degradation of formaldehyde (Kato et al., (1983), Attwood and Quayle (1984), Azachi et al., (1995), Doronia et al., (1997), Van Dijiken et al., (1975), Pilat and Prokop, (1976) where the concentration of formaldehyde was less than 400 mg/L and were conducted under non-saline conditions. In the present study, we have used a bacterial consortium to examine the growth and utilization of formaldehyde at different concentrations of NaCl (1%, 3%, 5% and 7%). At 100 mg/L of formaldehyde with 1% NaCl, the cfu/ ml in log phase showed 7 x 103, with the minimum removal efficiency of 31 %. At 3 % NaCl, the degradation was 87 % with a cell count of about 8 x 10 6cfu/mL and maximum degradation was obtained at 5 % NaCl (97 %) in 96 h which was shown by increased cell count of 7 x 107 without any lag phase. When the concentration of NaCl was increased to 7 %, the degradation efficiency decreased to 38% with decrease in the cell count to 6 x 104, which shows that the consortium could not mineralize formaldehyde effectively above 5 % NaCl. DM2 strain isolated from sea water could utilize formaldehyde (200mg/L) only with 3 % NaCl Yamazaki et al (2001). Azachi et al., (1997) reported the degradation of formaldehyde (75-100 mg/L) by Halomonas which grew at salt concentrations from zero to 20%. In the present study, degradation of formaldehyde was achieved only in the presence of NaCl, when the concentration of NaCl was 1 %, only minimal degradation (31%) was achieved, which proved that the consortium required salt for its growth on formaldehyde. Hence the bacterial strains in the consortium were moderately halophilic in nature.
Degradation of Different Concentrations of Formaldehyde
Previous experiment proved that the bacterial consortium was able to grow at an optimum salinity of 5 %. Hence to test its ability to grow at higher concentration of formaldehyde was investigated. At 200 mg/L of formaldehyde, the degradation was 82 % and growth was 7 x 106 within 96 h, when the concentration was increased to 300 mg/L, the degradation reduced to 75 % with cell count of 5 x 10 5 and further at 400 mg/L of formaldehyde only 62 % efficiency was observed with a cell count of 6 x104 (Figure 2). Previous report show that isolated Pseudomonas putida strain A2 utilized 400 mg/L of formaldehyde in 100h (Adroer et al., (1990)). Azachi et al., (1995) isolated Halomonas sp. strain MAC which used up to 75-100mg/L of formaldehyde. Lu and Hegemann (1998) reported that under 200 or 400 ppm of formaldehyde concentration, >90% of formaldehyde was degraded by incubation with anaerobic sludge for 20 d.
Yamazaki et al., (2001) isolated a formaldehyde-resistant bacterium from sea water (designated as DM-2), able to degrade 200 mg/L formaldehyde at 3 % NaCl (70 % of formaldehyde remained in the medium after 20h). They also reported that when the concentration of formaldehyde was increased to 400 mg/L, the removal efficiency reduced to 45 %.
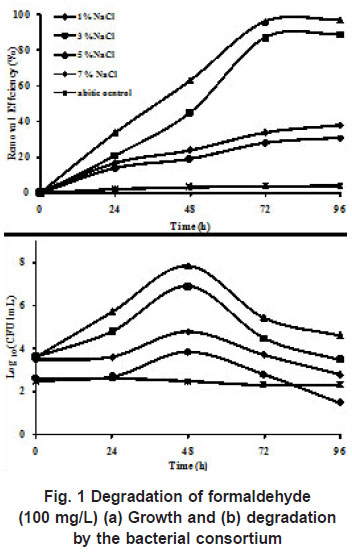 |
Figure 1: Degradation of formaldehyde
click here to view figure(100 mg/L) (a) Growth and (b) degradation by the bacterial consortium |
There are very few reports with individual strains degrading formaldehyde, where 400 mg/L concentration was used by cells of Pseudomonas putida in 100 h (Adroer et al., 1990). Halomonas DSM (7328) could utilize 75-100 mg/ L, (Azachi et al., 1995), E.coli (Gutheil et al., 1997), could not tolerate higher than 20 mg/L of formaldehyde. In our study, the bacterial consortium was able to remove 63 % of formaldehyde (400mg/L) in 96h under saline conditions.
Formaldehyde is metabolized either by the sequential action of formaldehyde dehydrogenase or formate dehydrogenase, or by using a cyclic pathway based on enzymes of the ribulose monophosphate cycle (Azachi et al., 1995). Hence future studies will be carried out on the enzymes involved in formaldehyde degradation under saline conditions.
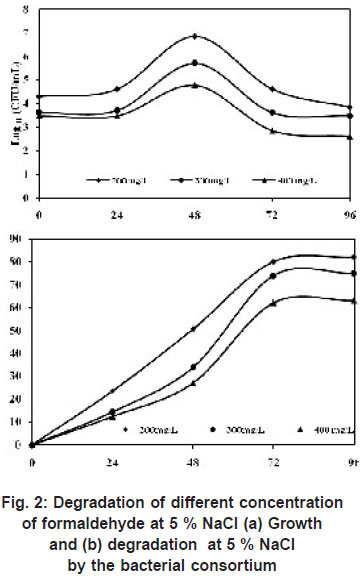 |
Figure 2: Degradation of different concentration of formaldehyde at 5 % NaCl (a) Growth and (b) degradation at 5% NaCl by the bacterial consortium click here to view figure |
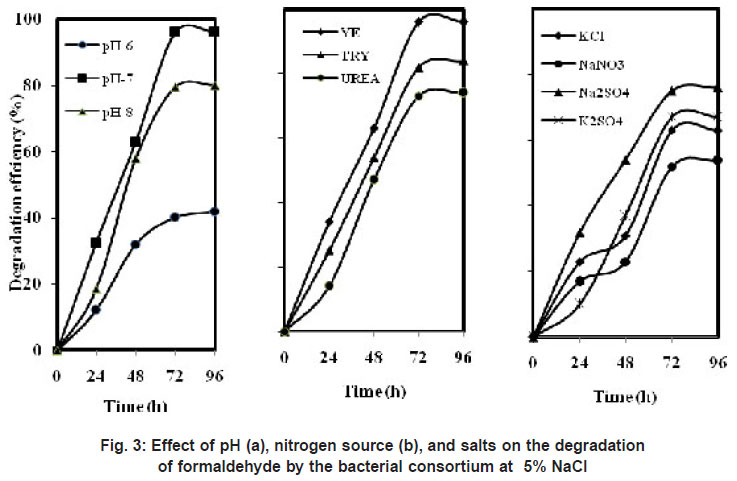 |
Figure 3: Effect of pH (a), nitrogen source (b), and salts on the degradation of formaldehyde by the bacterial consortium at 5% NaCl |
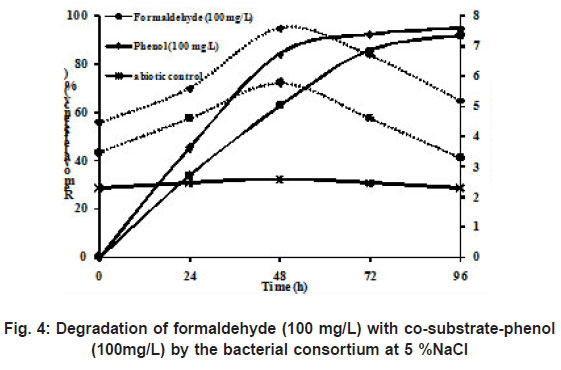 |
Figure 4: Degradation of formaldehyde (100 mg/L) with co-substrate-phenol(100mg/L) by the bacterial consortium at 5% NaCl |
Effect of pH, nitrogen sources and salts
The bacterial consortium could degrade formaldehyde at various NaCl concentrations ranging from 3 -7 %, but the optimum degradation of formaldehyde was achieved at 5 % NaCl at a concentration of 100mg/L. Hence the degradation of the formaldehyde (100mg/L) was evaluated for optimum pH, nitrogen source and co-substrate-phenol.
Effect of pH
The consortium could grow at pH from 6 to 8 (Figure 3 (a)). Highest degradation of 96 % was observed at 96 h at pH 7. About 80 % degradation was achieved at pH 8 and it further reduced to 42 % at pH 6.
Effect of Nitrogen Source
It was suggested that halophiles have more demanding nutritional requirements at high salt concentrations, and hence, complex media containing growth promoting factors may help to stimulate growth of halophilic bacteria at high salt concentrations (Oren et al., 2002). Our study also evaluated the effect of low concentrations of yeast extract, tryptone and urea (Figure 3 (b)). When yeast extract was used as nitrogen source maximum of 96 % degradation was observed and with tryptone 84 % and with urea 74 % as the nitrogen source. Overall, a maximum degradation of formaldehyde (100 mg/L) was obtained at 5 % NaCl and at pH-7.0 in the presence of 0.01% of yeast extract. Similar results were obtained by Carla et al., (2004) on the degradation of BTEX compounds under aerobic conditions by a halotolerant bacteria Marinobacter sp. which was able to degrade benzene within 8 days in the presence of yeast extract. An archaea (strain EH4) isolated from a salt marsh was able to degrade a higher percentage of eicosane (C20H42) in the presence of yeast extract, peptone, and Casamino Acids (Bertrand et al., 1994).
Effects of Salts
Industrial wastewater contains many other salts along with NaCl; hence the influence of different salts was studied under saline conditions in the degradation of formaldehyde. Figure 3 (c) shows the removal efficiency of formaldehyde with different salts. The isolated consortium was able to grow in the presence of different salts along with NaCl. About 76 % of formaldehyde was removed in the presence of Na2SO4 under saline conditions. With other salts KCl and K2SO4 the degradation of formaldehyde reduced to 63 % and 67 %. But the bacterial consortium showed least degradation of 54 % in the presence of NaNO3. This was in accordance with the results of Wang et al., (2009) where they studied the effects of salts on the degradation of phenol-cresol mixtures. Na2SO4 gave relatively higher removal efficiency of 90% than the other salts such as KCl and K2SO4.
Effect of Co-Substrate
Wastewater from phenol-resin manufacturing industry contains both phenol and formaldehyde as main components in the wastewater. Extensive literature about degradation of either phenol or formaldehyde has been reported under non-saline conditions. However, it is interesting to study the degradation of a mixture of phenol and formaldehyde under saline conditions. Hence, in this study an attempt has been made to examine the degradation of both phenol and formaldehyde simultaneously under saline conditions. During the study, the degradation of formaldehyde didn’t interfere with the degradation of phenol which is presented in Figure 4. The degradation of formaldehyde was 96 % at 100 mg/L, wherein the removal efficiency of phenol was 99 % with 100 mg/L of phenol. This is in agreement with the work of Eiroa et al., (2005) who showed that formaldehyde removal was not affected by the presence of phenol in the batch study under denitrifying conditions.
Conclusion
The present work has demonstrated the degradation of for maldehyde by the isolated consor tium at different salt concentrations, where the degradation was optimum at 5 % NaCl. The consortium could degrade up to 400 mg/L of formaldehyde, and mineralize phenol as the co substrate. To our knowledge this is the first report on the use of bacterial consortium, to degrade formaldehyde with phenol as the co substrate at different concentrations of NaCl. It is presumed that isolated bacterial consortium has potential application in remediation of wastewaters containing formaldehyde and phenol from high saline environments.
References
- Prado O. J., Eiroa M., Veiga M. C., Kennes C. Bioreactors for the treatment of industrial waste gases containing formaldehyde and other aliphatic compounds. In: Agathos, S.N., Reineke, W.
- (Eds.), Focus on Biotechnology, Biotechnology for the Environment: Wastewater Treatment and Modelling, Waste Gas Handling, Vol. 3C. Kluwer Academic Publishers, Dordrecht, The Netherlands, (2003) 259-273.
- Krick R. E., Othmer D. F. Encyclopedia of Chemical Technology, Vol 11, 3rd edn. Wiley, New York, 236-250 (1980).Bonastre, N., de Mas, C. and Sola, C., Vavilin equation in kinetic modelling of formaldehyde biodegradation. Biotechnology Bioengineering (1986) 28: 616-619
- Adroer N., Casas C., de Mass C., Sola C. Mechanism of Formaldehyde biodegradation by Pseudomonas putida. Applied Microbiology Biotechnology (1990) 33: 217-220.
- Kato N., Shirakawa K., Kobayashi H., Sakazawa C. The dismutation of aldehydes by a bacterial enzyme. Agricultural Biological Chemistry (1983) 47(1): 39-46.
- Yanase, H., Noda, H., Kiyotaka, A., Kita, K.and Kato, N., Bioscience Biotechnology Biochemistry. (1995) 59: 197-202.
- Gutheil, W.G., Kasimoglu, E., and Nicholson,P.C., Induction of glutathione-dependent
- Formaldehyde dehydrogenase activity in Escherichia coli and Haemophilus influenzae Biochemistry Biophysics Research Communication (1997) 238: .693-696.
- Attwood, M.M., and Quayle J.R.,Formaldehyde as a central intermediary metabolite of methylotrophic metabolism. In:Microbial growth on C1 compounds, R.L.Crawford, R.H. Hanson (Ed.), American Society for Microbiology, Washington, D.C (1984) 315-323.
- Kaszycki, P. and Ko³oczek H., Formaldehyde and methanol biodegradation with the methylotrophic yeast Hansenula polymorpha in a model wastewater system. Microbial Research (2000) 154: 289-296.
- Kaneko, Y., Ito, M., Ogura, Y., and Ishikawa,J., US patent no. (1985) 4: 505,821.
- Azachi, M., Henis, Y., Oren, A., Gurevich, P. and Sarig, S., Transformation of Formaldehyde by a Halomonas sp. Canadian Journal of Microbiology (1995) 41: 548-553.
- Yamazaki, T., Tsugawa, W. and Sode, K., Biodegradation of Formaldehyde by a Formaldehyde-resistant bacterium isolated from seawater. Applied Biochemistry Biotechnology (2001) 91-93: 213-217.
- Hidalgo, A., Lopategi, A., Prieto, M., Serra, J.L. and Llama, M.J., Formaldehyde removal in synthetic and industrial wastewater by Rhodococcus erythropolis UPV-1. Applied Microbiology Biotechnology (2002) 58: 260-263
- Cvetkovic, Z., Suhadolnik, Z., Grilc, V., Recelj,T., Golob, J., Vrtovsek, J., Biological treatment of waste-water from a phenolformaldehyde resins production. Chemical and Biochemical Engineering Q. (1988) 2: 262-268.
- Vidal, G., Jiang Z.P., Omil, F., Thalasso, F.,Mendez, R.M. and Lema, J.M., Continuous anaerobic treatment of wastewaters containing formaldehyde and urea Bioresource Technology (1999) 70: 283-291.
- Alva, V.A. & Peyton, B.M., Phenol and catechol biodegradation by the haloalkaliphile Halomonas campisalis: influence of pH and salinity, Environmental Science and Technology (2003) 37: 4397-4402.
- Nash, T., The colorimetric estimation of Formaldehyde by means of the hantzsch reaction. Journal of Biochemistry, (1953) 55: 416-421.
- Doronia, N.V., Ezhov, V.A. & Trotsenko, Yu.A.,Aerobic biodegradation of Formaldehyde,methanol and methylamine by immobilized Methylobacterium extorquens cells. Applied Biochemistry Microbiology (1997) 33: 138-141.
- Van Dijiken, J.P., Veenhuis, M., Kreger-VanRij, N.Y.W. & Horder, W., Microbodies in methanol-assimilating yeasts Archieves Microbiology. (1975) 102: 41-44.
- Pilat, P. and Prokop, Application of mathematical optimization methods in Microbiology Foila Microbiology (1976) 21: 306-314.
- Lu, Z. and Hegemann, W., Anaerobic toxicity and biodegradation of formaldehyde in batch cultures Water Research (1998) 32(1): 209-219.
- Oren, A., Gurevich, P., Azachi, M. & Henis,Y., Microbial degradation of pollutants at high salt concentrations, Biodegradation (1992) 3: 387-398.
- Carla, A. N. and Babu, Z. F., Biodegradation of benzene by halophilic and halotlerant bacteria under aerobic conditions. Applied Environmental Microbiology (2004) 70(2):1222-1225.
- Bertrand, J. C., Almallah, M. Acquaviva, M.and G. Mille., Biodegradation of hydrocarbons by an extremely halophilic archaebacterium. Letters in Applied Microbiology, (1990) 11: 260-263.
- Wang, P., Qu Yuanyuan and Zhou J.,Biodegradation of Mixed Phenolic Compounds Under High Salt Conditions and Salinity Fluctuations by Arthrobacter sp. W1 Applied Biochemistry Biotechnology (2009) 159(3):623-33.
- Eiroa, M., Vilar, A., Amor, L., Kennes, C. &Veiga, M.C., Biodegradation and effect of Formaldehyde and phenol on the denitrification process. Water Research (2005) 39:449-455.






