Isolation and Characterisation of Diazotrophic Bacteria from Rhizosphere of Different Rice Cultivars of South Assam, India
Folguni Laskar1 * and G.D. Sharma2
1
Department of Life Science and Bio-Informatics,
Assam University,
Silchar,
788 011
India
2
Department of Life Sciences,
Bilaspur University,
Chattisgarh,
495 009
India
DOI: http://dx.doi.org/10.12944/CWE.8.1.20
Copy the following to cite this article:
Laskar F, Sharma G. D. B. Isolation and Characterisation of Diazotrophic Bacteria from Rhizosphere of Different Rice Cultivars of South Assam, India. Curr World Environ 2013;8(1) DOI:http://dx.doi.org/10.12944/CWE.8.1.20
Copy the following to cite this URL:
Laskar F, Sharma G. D. B. Isolation and Characterisation of Diazotrophic Bacteria from Rhizosphere of Different Rice Cultivars of South Assam, India. Curr World Environ 2013;8(1). Available from: http://www.cwejournal.org/?p=3468
Download article (pdf) Citation Manager Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2013-03-24 |
|---|---|
| Accepted: | 2013-04-14 |
Free living nitrogen fixing bacteria have been considered as an alternative for inorganic nitrogen fertiliser for promoting plant growth (Ladha and Reddy, 2000; Park et al., 2004). Inspite of the fact that a variety of nitrogen fixing bacteria have been isolated from the rhizosphere of various crops, interest in isolating more beneficial plant growth promoting bacteria has increased recently due to their potential use as bio fertiliser (Vessey, 2003). Cultivated rice (Oryzasativa) originated from species of wild rice and was domesticated several thousand years ago (Hoshikawa,1989; Oka,1988; Kennedy et al., 2001). Wild rice species are likely to harbour unique populations of nitrogen-fixing bacteria that differ from those in extensively bred modern varieties of cultivated rice (Hurek et al., 2000). Production of phytohormones (Tienet al., 1979; Haahtelaet al., 1990) and competitive suppression of plant phytopathogens (Glick, 1995, Park et al., 2004) are the beneficial effects of these microbes on plant. The agricultural soil of South Assam experience heavy annual rainfall, resulting in leaching of nutrients causing economic loss to the farmers (Anonymous, 2007). The use of bio-fertilisers may be an alternative to the chemical fertiliser for achieving sustainable rice farming while improving productivity in rice-agro ecosystem.
The objective of the present study was to isolate, identify and molecular characterisation of diazotrophic bacteria from the rhizosphere of native local varieties of rice from different microhabitats of South Assam, India and characterise them for nitrogen fixation and plant growth promoting activities.
Materials and Methods
Bacterial Growth Media
Burk’s N-free medium comprising : 10g glucose, 0.41g KH2PO4, 0.52g Na2SO4, 0.2g CaCl2, 0.1g MgSO4.7H2O, 0.005g FeSO4.7H2O, 0.0025g Na2MoO4.2H2O, 1.8g agar (all per litre distilled water) for semi solid and 15g agar for solid medium was used throughout the study (Wilson and Knight, 1952). The pH of the medium was adjusted to 7±0.1 before autoclaving at 1210C for 15mins
Soil Sample Collection and Isolation of Diazotrophs
Ten native rice cultivars grown under different microhabitats in Karimganj districts of South Assam were randomly selected. Riceplant were uprooted from a depth of 0-5cm for rhizosphericsoil sampling. The samples were then immediately placed in sterilized plastic packs and sealed brought to the laboratory and stored at a temperature of 4oC. Ten grams of the rhizosphericsoil sample was transferred to a 250ml of Erlenmeyer flask containing 90ml of sterile distilled water and shaken (120rpm) for 30mins. Serial dilution was made and 0.1ml aliquots (103-105) were spread on plates containing Burk’s N-free medium. The plates were incubated for 7 days at 300C. Morphologically different colonies appearing on the medium were isolated and sub-cultured for further analysis.
 |
Figure 1: Phylogenetic tree derived from analysis of the 16S rDNA sequences of Strain KR-23, KR-4, KR-6, KR-16 and KR-7 and related sequences obtained from NCBI. Scale bar, 0.02 substitutions per nucleotide position Click here to View figure |
Phenotypic Characterisation of the Bacterial Isolates
Physiological and biochemical characters of the bacterial isolates were examined according to the methods described in Bergey’s Manual of Systematic Bacteriology (Holt et al., 1994). Colour, pigment, form, diameter, elevation, margin, surface, opacity and texture of the colonies of respective isolates were observed. Motility and morphology were examined with the help of phase contrast microscopy. The gram reaction was performed as per standard protocol. Plate assays were performed for determining the starch hydrolysis, carbon source utilisation of the isolates. MR-VP test, indole test, urea hydrolysis test were also performed.
16S rDNA Gene Amplification and Sequencing
Genomic DNA was obtained by using standard bacterial procedure (Sambrook et al., 1989; Park et al.,2004). Extraction of the lysate was done two times with chloroform to remove residual phenol. The primers used for PCR amplification of the 16s ribosomal DNA are 27F (5∕-AGAGTTTGATCCTGGCTCAG-3∕) and 1492R (5∕-TACGGTTACCTTGTTACGACTT-3∕) (Weisberg et al.,1999). Each PCR mixture (50µl) contained primers (at a concentration of 20pmol) a mixture of dNTP (Promega Co., Southampton, England) (at a concentration of 200µM), Taq polymerase buffer and chromosomal DNA (ca 100ng) and enzyme (4µl) were added to the reaction mixture. The thermo cycling condition consisted of an initial denaturation step at 940C for 4min, 35 amplification cycles of 940C for 1 min, 580C for 1 min and 720C for 3 min and a final extension step of 720C for 10 min with Gene-Amp PCR system (Perkin-Elmer Co., Norwalk, Conn.). PCR products were run and visualised on a 0.7% agarose gel. The 16S rDNA nucleotide sequences was determined by PCR direct sequencing using ABI PRISM 310 Genetic Analyser (PE Applied Biosystems, Foster City, CA) and Big Dye Terminator cycle system (PE Applied Biosystems). Related sequences were obtained from Genebank Database, National Centre for Biotechnology Information(NCBI) using BLAST, Version 2 (Altschul et al., 1990; Park et al.,2004).The sequences were aligned and the consensus sequences was computed using Clustal W software. The evolutionary history was inferred using the Neighbor-Joining method (Saitou N et al., 1987) and the evolutionary distances were computed using the Maximum Composite Likelihood method (Kumar S et al., 2004) MEGA version 5 (Kumar et al., 1993).
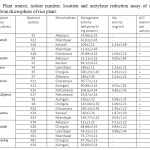 |
Table 1: Plant source, isolate number, location and acetylene reduction assay of different isolates from rhizosphere of rice plant. Click here to View table |
Acetylene Reduction Assay (ARA)
Nitrogenase activity of bacterial strains was determined in semisolid nitrogen free malate medium (by using acetylene reduction assay (ARA; HARDY et al. 1973). Pure bacterial colonies were inoculated on to NFM medium in McCartine vials of 10 ml capacity and incubated at 28 ± 2 °C for 48 h. Acetylene gas (10% vol/vol) was injected to the vials. After incubation for 16 h at 28 ± 2 °C, gas samples (100 μl) were analyzed on a Hewlett Packard gas chromatograph (Model HP 6890, USA) using Porapak-N column and H2 flame ionization detector. After completion of ARA the cell were predigested by adding 10% SDS and sonicated briefly. Protein concentration in the resulting mixture of suspension was determined by Lowry’s method. (Lowry et al.,1951).
Indole-3-Acetic Acid Production
IAA production by the isolates was determined by growing the isolates in Burk’s medium supplemented with L-tryptophan (100mg/l) at 300C. The supernatant of the culture fluid was obtained by centrifuging the stationary phase cultures at 10,000rpm for 15min and the pH was adjusted to 2.8 with 1N HCL. The auxins from the acidified cultures extracted with the equal volumes of ethyl acetate (Park et al., 2004; Tienet al., 1979) was evaporated to dryness and re-suspended in 4ml of ethanol. Analysis of the samples was done by HPLC (Water series 474, USA) using UV detector on a Nova-Pak 5-ODS C-18 column. Methanol: acetic acid: water (30:1:70 v/v/v) was used as mobile phase at a rate of 0.6mlmin-1 (Rasulet al., 1998). Pure indole-3-acetic acid (Sigma USA) was used as standard. The IAA of the samples was quantified by comparing the retention times, peak areas, and UV absorbance spectra with those of the standard using a Millenium 32 Login software with an interface (Waters, USA) attached with computer.
Phosphorous Solubilisation
Test for P-solubilisation was done following Goldstein (1986). The appearance of clearing zone around bacterial colonies after 96h of growth at 300C was used as indicator for P-solubilisation. Plates inoculated with heat killed cells served as control.
ACC Deaminase Activity
ACC deaminase activity was determined following the method described by Glick et al., 1995. For this 1 µl of each LB pure bacterial culture was inoculated into agar plates containing NFb or NFb-ACC modified by addition of 1-aminocyclopropane-1-carboxylate (5.0 gl-1) as unique nitrogen source. Plates were incubated at 280C and observed daily for colony formation for upto 4 days. Colonies were re-inoculated and incubated in the same experimental conditions. Newly colonies formed in NFb with addition of ACC were considered positive for ACC deaminase activity.
Statistical Analysis
Results of the measurements were subjected to analysis of variance (ANOVA) using SAS package, Version 8.2 (SAS, 2001).
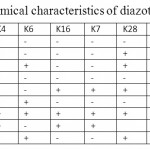 |
Table 2: Biochemical characteristics of diazotrophic isolates Click here to View table |
Results
The isolation and identification of diverse groups of diazotrophs has paved the way to decrease the costs and use of inorganic N-fertiliser as well as minimises the risk of pollution of agro-ecosystem from the application of chemical fertilisers. Previous researches on isolation of nitrogen fixing bacteria have revealed a broad diversity of diversity of diazotrophs associated with different crop rhizosphere (Vessey, 2003). Diverse free living or associative N2-fixing microorganisms (aerobes, facultative anaerobes, heterotrophs, phototrophs) grew in wetland rice fields and contributed to soil N. The location, variety of rice plant, soil type and ARA, IAA activity and ACC deaminase activity of different isolates from rhizosphere soil has beenshown in Table 1. Total 25 free living nitrogen fixing bacterial isolates were isolated on Burk’s nitrogen free media. The isolates were then purified and sub-cultured on solid nitrogen free medium.
The ability to reduce acetylene was an indicator of nitrogen fixing potential and was specific for monitoring functional nitrogen fixing potential (Andrade et al., 1997). Out of 25 isolates, 11 were selected for their efficiency in nitrogen fixing, having ARA activity , IAA activity and ACC deaminase activity. Among all the strains KR-23 exhibited highest nitrogenise activity (300 nmolC2H4 h-1mg-1protein).
Biochemical characterisation of the strains showed that all the strains were gram –ive, motile rods and having varying metabolic activities. The biochemical characteristics of all the isolates are given in Table 2.
From 16S rDNA sequence analysis of the strains it was observed that ninety nine percent sequence identity was observed between the 16S rDNA sequence of KR-4 with Pseudomonas putida; KR-23 and KR-5 with Spingomonasazotifigens; KR-6 with Stenotrophomonasmaltophila; KR-16 with Herbispirillum sp. KR-7 with Herbispirillumrubrisubalbicans, KR-28 with Klebsiella pneumonia, K15 with Pantoeaagglomerans, KR-34 with Acinetobacterradioresistance, KR-18 with Enterobactercloaceaesubspdissolvens, KR-8 with Alcaligenesfaecalis and KR-20 with Achromobacterxyloxidans. Isolation of all the above bacteria were already reported from different crops by many workers (Bhromsiriet al., 2010; Sa et al. 2009; Uretaet al., 1995; Gholamiet al., 2009; Xieet al., 2006). A phenogram reflecting the relationship among the strains and candidate sequences of various nitrogen fixing strains obtained from database of NCBI has been presented in Fig 1. The sequences obtained were submitted to NCBI with accession numbers KR-4: JN222977, KR-23: JN085438, KR-5: JN085437, KR-6: JN085439, KR-16:JF990839, KR-7:JF906701, KR-28:JN162393, KR-8: JN162397, KR-20: JN162396, KR-15: JN162392; KR-34: JN162392, KR-18: JN162395.
Discussion
Plant growth promoting rhizobacteria offer an environmentally sustainable approach to increase crop production and health. The IAA production and ACC deaminase activity of the microbes help in the stimulation of growth and pathogenesis of the plants. In this study all the 11 strains produced considerable amount of IAA and are also capable of ACC deaminase activity which can be comparable to the earlier studies on various bacteria by different workers (Malik et al., 1997; Sucksstorff and Berg, 2003; Park et al., 2004; Xieet al., 2006). The amount of ARA and IAA activity and the capability of ACC deaminase activity is shown in Table 1. Among all the isolates Sphingomonasazotifigens, Pseudomonas putida, Pantoeaagglomerans, Enterobactercloaceae subsp. dissolvens, Klebsiellapneumoneae and Herbispirillumsp can solubilise phosphorous.
This study reports the isolation and characterisation of the strains of S.azotifigens, S. maltophila, P. putida, Herbispirillum sp., H.rubrisubalbicans,P.agglomerans, A.xyloxidans, A radioresistance, A. faecalis, E. cloaceae subsp. dissolvens and K. Pneumoneae from the inorganic fertiliser rich rhizosphere soil of rice agro-ecosystem of South Assam confirming their nitrogen fixing potential.
Sphingomonasazotifigens is reported from the rice fields of South Assam for the first time in India in our study. All the 11 isolates are capable of IAA production and ACC deaminaseactivity, among them some are capable of P-solubilisation. The isolates Pseudomonas putida, Sphingomonasazotifigens, Herbispirillum sp and has been regarded as PGPR in earlier studies (Gholami, 2009; Park et al., 2004; Baldaniet al., 2009), but Stenotrophomonasmaltophila and Herbispirillumrubrisubalbicans, as producer of IAA and capable of ACC deaminase is also reported first time in our study. These two bacteria are reported as pathogenic and endophytes in many earlier study (Reinhardt et al.,2008;Denton et al.1999; Hale et al., 1972; Gillis et al., 1990). An increased knowledge of the role of these isolates in plant growth promotion under pot culture as well as field condition is required to prove them as plant growth promoting rhizobacteria.
Acknowledgement
The authors are grateful to the Head, Department of Life Science, Assam University (Silchar), India for providing laboratory facile.
Reference
- Altschul S.F., Gish W., Miller W., Myers E.W. and Lipman D.J., J. Mol. Biol., 215, 403 (1990).
- Andrade G., Esteban E., Velascol L., Maria J.L. and Bedmar E.J., Plant Soil 197, 19 (1997), http://dx.doi.org/10.1023/A:1004211909641
- Baldani I.J. and Baldani L.V., Anais Da Acad. Brasiliera De Ciencias Sci., 77, 549 (2005), http://dx.doi.org/10.1590/S0001-37652005000300014
- Bhromsiri C. and Bhromsiri A ., Thail J. Agric. Sci., 43(4), 239 (2010).
- Cheng-Hui Xie and Yokota A., International Journal of Systematic and Evolutionary Microbiology, 56, 889 (2006), http://dx.doi.org/10.1099/ijs.0.64056-0
- Denton M. and Kerr K.G., Clin.Microbiol. Rev., 11, 57 (1998).
- Reinhardt E. L., Ramos P.L., Manfio G.P., Barbosa H.R., Pavan C. and Carlos A. Moreira-Filho C.A., Brazilian Journal of Microbiology, 39, 414 (2008), http://dx.doi.org/10.1590/S1517-83822008000300002
- Gholami A., Shahsavani S. and Nezarat S., World Academy of Science, Engineering and Technology 49 (2009).
- Gillis M., Dobereiner J., Pot B., Goor M., Falsen E., Hoste B., Reinhold B. and Kersters K. In M. Polsinelli, R. Materassi, and M. Vin-cenzini (ed.), Nitrogen fixation. Kluwer Academic Publisher, Dordrecht, TheNetherlands. 293 (1990).
- Glick B.R., Can. J. Microbiol. 41, 109 (1995), http://dx.doi.org/10.1139/m95-015
- Goldstein A.H., Am. J. Alter. Agric., 1, 51 (1986).
- Gyaneshwar P., James E.K., Mathan N., Reddy P.M, Reinhold B., Hurek and Ladha. J. Bacteriol.,183, 2634 (2001), http://dx.doi.org/10.1128/JB.183.8.2634-2645.2001
- Haahtela K., Konkoo R., Laakso T., Williams P.H. and Korhonem T.K., Mol. Plant Microbe Interact., 3, 358 (1990), http://dx.doi.org/10.1094/MPMI-3-358
- Hale C. N. and Wilke J.P., J. Agric. Res., 15448 (1972).
- Hardy R.W.F., Holsten R.D. and Jackson E.K., Plant Physiol., 43, 118 (1968).
- Holt J.G., Kreig N.R., Sneath P.H.A., Staley J.T., Williams S.T., Williams and Wilkins, Baltimore, USA (1994).
- Hoshikawa, K. Anthesis, Science of the Rice Plant, Volume One. Morphology, 339. Tokyo: Food and Agriculture Policy Research Center (1993).
- Kennedy I. R., Choudhury A.T.M.A. and Kecskés M.L., Journal of Soil Biology and Biochemistry, 36(8), 1229 (2004), http://dx.doi.org/10.1016/j.soilbio.2004.04.006
- Ladha, J.K. and Reddy, P.M., The quest for nitrogen fixation in rice. Proceedings of the Third Working Group Meeting on Assessing Opportunities for Nitrogen Fixation in Rice, 9–12 Aug. 1999, IRRI, Los Banos, Laguna, Philippines, 354 (2000).
- Lowry H.O., Rosebrough N.J., Farr A.G. and Randall R.J., J. Biol. Chem., 193, 265 (1951) .
- Park M., Kim C., Yang J., Lee H., Shin W, Kim S. and Sa T., Microbiological Research 160, 127 (2005), http://dx.doi.org/10.1016/j.micres.2004.10.003
- Oka, H.I.. Origin of Cultivated Rice. Tokyo: Japanese Scientific Societies Press (1988).
- Rasul G., Mirza M.S., Latif F. and Malik K.A., Kluwer Academic Publishers, Dordrecht, The Netherlands, 25 (1998).
- SAS, Institute Inc., SAS user’s guide, Version 8.2. SAS Institute Inc., Cary, North Carolina, USA (2001).
- Saitou N. and Nei M., Mol. Biol. Evol., 4, 406 (1987).
- Shahi S.K., Rai A.K., Tyagi M.B., Sinha R.P. and Kumar A., African Journal of Biotechnology, 10(42), 8296, (2011).
- Suckstorff I., Berg G., J. Appl. Microbiol., 95, 656 (2003), http://dx.doi.org/10.1046/j.1365-2672.2003.02021.x
- Tien T.M., Gaskins M.H. and Hubbel D.H., Appl. Environ. Microbiol., 37, 1016 (1979).
- Ureta A., Alvarez B., Ramon A., Vera M A. and Martinez-Drets G., Plant Soil 172, 271 (1995), http://dx.doi.org/10.1007/BF00011329
- Vessey J.K., Plant Soil 255, 571 (2003), http://dx.doi.org/10.1023/A:1026037216893
- Weisburg W.G., Barns S.M., Pelletier D.A.and Lane D.J., J. Bacteriol. 173, 697 (1991).
- Wilson P.W. and Knight S.C., Burguess, Minneapolis, USA, 49 (1952).







