The Presence of Mercury Resistant Bacteria in Sediment of Gold Processing Plant at Waekerta Village of Buru District, Maluku Province and their Activity in Reducing Mercury
Sarmawaty Kotala1 * , Retno Kawuri2 and Ida Bagus Wayan Gunam3
DOI: http://dx.doi.org/10.12944/CWE.9.2.07
Mercury was one of the heavy metal polute in environment and had the toxic characteristic to the living creatures. Golden mining in Waeapo subdistrict used mercury to extract the gold and exile the waste to the environment freely. Several precedented research showed that waste sediment of gold processing contains mercury resistance bacteria. Mercury resistance bacteria can be used as bioremediation agent because those bacteria can reduce mercury. Mercury resistance bacteria has mer operon which contained in plasmid. The goal of this research is to isolate mercury resistance bacteria which is able to grow on medium nutrient agar (NA) containing 500 ppm of HgCl2 and to analyze the capability in HgCl2 reduction in nutrient broth (NB) medium. Bacteria isolation was done by platting method on Nutrient Agar containing 10 ppm of HgCl2. Bacteria identification was done by kit Microgen TM GnA + B-ID System and to know bacteria capability in reducing mercury was done by CV-AAS (Cold Vapour Atomic Absorption Spectrophotometer). Result showed, that the bacteria found in this research were Bacillus sp and Aeromonas hydrophila. Both of these bacteria were able to reduce HgCl2 in the amount of 98,7% for Bacillus sp and 98,33% for Aeromonas hydrophila. In the future those bacteria can be use as bioremediation agent.
Copy the following to cite this article:
Kotala S, Kawuri R, Gunam I. B. W. The Presence of Mercury Resistant Bacteria in Sediment of Gold Processing Plant at Waekerta Village of Buru District, Maluku Province and their Activity in Reducing Mercury. Curr World Environ 2014;9 (2) DOI:http://dx.doi.org/10.12944/CWE.9.2.07
Copy the following to cite this URL:
Kotala S, Kawuri R, Gunam I. B. W. The Presence of Mercury Resistant Bacteria in Sediment of Gold Processing Plant at Waekerta Village of Buru District, Maluku Province and their Activity in Reducing Mercury. Curr World Environ 2014;9(2). Available from: http://cwejournal.org?p=502/
Download article (pdf) Citation Manager Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2014-04-21 |
|---|---|
| Accepted: | 2014-05-17 |
Introduction
Mercury utilizing in golden mining could produce waste, which contains mercury and causes environment pollution. Mercury belongs to heavy metal which is toxic to living creatures. Mercury can attack the arrangement of central nervous and causes memory loss, tremors and decreases motion capability. Poisoning causing destruction of a fetus has been detected. Minimata desease in Japan is the example of mercury poisoning.1,2
Mercury as a pollutant in the environment need attention and problem solving. Mercurydetoxificationcan be donechemicallybyprecipitation, coagulation, reverseosmosis, ionexchangeresinsandadsorptionusingactivated carbon.3,4 However, thisprocessis relativelyexpensiveandcould causenew problems, namelythe accumulation ofthese compoundsinsedimentandaquatic organisms.4
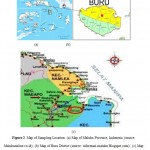
Mercury detoxification can be done by using mercury resistance bacteria which have mercury resistance gen, called mer operon,.4,5 There is gold mining at Waekerta village, Subdistrict Waeapo, Maluku Province, Indonesia,where theprocessing ofgoldusingmercuryandthe wasteis discharged intothe environmentwithout regard tothecontaminationoccurred(Figure 1).Based on this background, it is necessary to isolate mercury resistance bacteria from sediment of gold processing which able to grow on NA medium containing 500 ppm of HgCl2 and to analize the capability of reduction of HgCl2.
|
|
Figure 1: Gold mining in Waekerta village (a) Gold |
Material and Methods
Research Materials
This study used asample ofsedimenttakenfroma waste disposal siteof gold processing in Waekerta Village, Maluku. Materials usedin this study were Nutrient Agar (Merck),Nutrient Broth (Merck), and HgCl2.
Research Instrument
The instrument used in this research is a hot plate, autoclave, vortex, incubator, kit Microgen TM GnA + B-ID System, microscopes, spectrophotometers, CV-AAS, and laminar air flow cabinet.
Sampling
Samples were takenas much as 20% from42gold processingsites inWaekerta Village, so that9 locations were choose to get the sediment samples. Samples taken from each location on 5 different points were then mixed into one. Land sample from mining land was used as comparative so that total of all samples become ten.Locationof sampling sitesin the villageof Waekertaas shown in Figure2.
|
|
Figure 2: Map of Sampling Location: |
Bacteria Isolation
Mercury resistance bacteria isolation was done by spread plate method.6 Sedimen and soil sample were diluted in a series(10-1, 10-2and10-3) with saline solution(0.85% NaCl). From the 10-3 dilution were taken0,1 ml and spread on petri dishes containing selective media namely nutrient agar (NA) containing10ppm of HgCl2. Then incubatedat room temperaturefor 3 days. Grown bacterial isolates with different colonies morphological characterswere reisolated aga into a new mediumin order to get pure cultures and stored in an agar slant for further testing.
Mercury Recistance Bacteria Selections
Bacteria selection is based on the ability of bacteria lisolatesgro wninmedium with various HgCl2 concentrations. Bacterial isolateswere grownbystreaking method on NA medium which contain 25ppmof HgCl2 and incubatedat room temperature for 24hours. If the isolatesgrow, then chese bacteria lisolates were re-grown by streaking method on the Name diumadded with HgCl2 with a higher concentration of 50ppm, 100ppm, 250ppm, 400ppm, 500ppmin order to obtain superior isolates,which were able to live in the highest HgCl2 concentration. Purified isolates was storedin nutrient agar slant medium with a temperature of-20°C.
Mercury Resistance Bacteria Identification
Parameters observed for identification of mercury-resistant bacteria are colony form on NA medium, Gramstaining, andcharacterphysiology (biochemical test). Physiological characteristicswere tested using Microgen™ kitGNA+B-ID System Identification (Microgen Bioproduct, UK).
Determination of Optimum Temperature on the Growth of Mercury Resistant Bacteria
To determine the optimum growth temperature, the bacterial isolates were grown on nutrient broth medium and incubated a variety of temperature is: 25°C, 30°C, 37°C, and 45°C. Cultures were incubated at this temperature for 24 hours. Further growth of the isolates was measured degree of turbidity with a spectrophotometer at a wavelength of 620 nm. Absorbance values of bacterial cells can be observed at a wavelength of 620 nm, each treatment was repeated 3 times.
Determination of Optimum pH on Mercury Resistant Bacteria Growth
To determine the optimum pH of growth, the bacteria lisolates were grown innurient broth with a pH of 5, 6, 7, 8, and 9. Cultures were incubatedat the optimum temperature for 24 hours. Growth of isolates was measured with a spectro photo meter at a wavelength of 620 nmand each treatment was repeated 3 times.
Determination of Bacterial Growth Curve
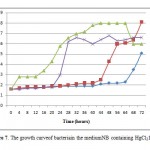
Aseptically one ose of superior bacterial isolates at the age of 24 hours (isolates were rejuvenated on NA medium containing 10 ppm of HgCl2) was inoculated in 100 ml of NB medium in erlenmeyer flask, incubated at room temperature on a rotary shaker (100 rpm). 24-hour-old culture was washed using saline solution, then 5ml of the culture was taken and inoculated into 45 ml of NB mediumat a concentration of 10 pp mHgCl2 andin cubatedat room temperature on arotary shaker(100 rpm). Suspension culture absorbance values were measured at a wave length of 620 nm. Absorbance measurements were started from 0 hour up to 72 hours with an interval of 4 hours. Obtained absorbance data was then conversed into the growth curve. On the x-axis is time and and on the y-axis is absorbance. The growth curve will be compared with the growth curve of bacteria in NB medium without HgCl2.
MercuryReducing BacteriaActivity Test
Mercury reducing bacteria activity test was carried out to look at the ability of superior isolates inreducing Hg. In this testing phase bacterial isolates were grown in NB medium for 24 hours in 250 ml erlenmeyer, then isolated cells were washed using saline solution and the absorbance was measured using a spectrophotometer at a wavelength of 620 nm. Culture with absorbance value of 2 was taken 0.1 ml and grown in 50 ml NB medium containing a concentration of 100 ppm HgCl2,then incubated for 7 days on top shaker (100 rpm). Furthermore, bacterial cells were separated from the medium by using a membran filter with the size of 0.2 µm. Hg concentration remaining in the medium was measured by Cold Vapour NB Atomic Absorption Spectrophotometer (CV - AAS). In addition, NB medium containing 100 ppm of HgCl2 without inoculated with bacteria resistant to mercury was used as a positive control and NB medium without HgCl2 and mercury resistant bacteria was used as negative control. The principle of CV-AAS working is to change the mercury dioxide compounds into the mercury ion, mercury ion subsequently reduced to metallic mercury and the cold vapor atomic absorption of it was analyzed at a wavelength of 253.7 nm. Reagents used were SnCl2 reductant, H2SO4 + HCl acid solution ( Rondonuwu, 2011).To determine the levels of mercury removal efficiency, this formula was used
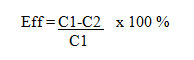
Whereas: C1 = First concentration (ppm);C2 = Final concentration (ppm); Eff = Efficiency
Data Analysis
The data were analyzed qualitatively and quantitatively. Qualitatively is by describing the results of the characterization and identification of mercury-resistant bacterial isolates were able to reduce mercury. Quantitatively, on the pH test and growth curve measurement was done by measuring the number of bacterial cells through the absorbance. The data obtained was made in the form of a bar graph, but the growth curve in the form of a line graph using Microsoft Excel program.
Results and Discussion
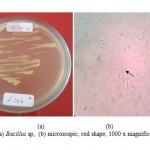
Two isolates of mercury resistant bacteria capable of living NA medium containing 500 ppmof HgCl2 was found ingold processing sediment samples. The isolates were L.10bandL.10c. After identification, these isolates were identified as Bacillusspand Aeromonashydrophyla. Both macroscopic and microscopic forms of the isolatescan be seenin Figure3 and Figure 4 as well as the character of each isolate are shown in Table1.
|
|
Tabel 1: Characteristic of L.10b and L.10c |
|
|
Figure 3: (a) Bacillus sp, (b) microscopic, |
|
|
Figure 4: (a) Aeromonas hydrophila, (b) microscopic, |
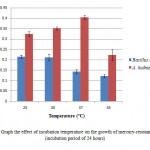
Bacillussp was found as a mercury resistant bacteria in Japan and India.7 In addition, Bacillussp was also found in the Tondano river, Indonesia.8 Bacillus cereusandBacillussubtilis found in the Kalimas river Surabaya are also resistant tomercury.9 Bacillusspis moreoften found as mercury resistant bacteria compared to Aeromonashydrophila. Aeromonashydrophyla has been found in gold mining sediment contaminated by HginBandung, West Javaandable to grow at 550mg/LHgCl2.10 In addition, some strains of A.hydrophilais found in seawater, fish, and waste water contaminated by heavy metalsinTunisia.11 Temperature is one of the environmental factors that influence the growth of bacteria. Bacillus sp. has the highest absorbance value (0.216) at 25ËšC and the lowest (0.118) at a temperature of 45ËšC. Aeromonas hydropila has a high absorbance value (0.404) at 37ËšC and the lowest (0.224) at a temperature of 45ËšC (Figure 5). Temperature effect on bacterial growth because temperature affects the activity of enzymes in metabolism. The temperature affect the chemical reactions in the process of bacterial growth, growth rate, and the total amount of the growth of micro organisms.12 Although the absorbance values of different bacteria are categorized both mesophilic bacteria. Mesophilic bacteria is a group of bacteria that can grow at a temperature of 20-45ËšC.13
|
|
Figure 5: Graph the effect of incubation |
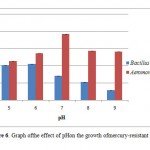
Bacterial growth can be affected by various environmental factors, one of which is the pH of the medium. The degree of acidity of the medium affects the growth of Bacillus sp and A. hydro phyla. Bacillus sp grows optimally at pH 6 with a absorbance value of 0.106 and the absorbance values decreased when the pH of the medium increased (Figure 6).
|
|
Figure 6: Graph ofthe effect of pHon the |
In contrast to Bacillus sp,A. hydrophila has the highest absorbance value (0.192 ) at pH 7 and the lowest (0.11) at pH 5 (Figure 6). The degree of acidity affects the growth of bacteria because the pH affects the enzymes in the metabolism of bacteria. Enzyme activity will decrease if the pH is not appropriate, this is because the enzyme will be active in a proper state of ionization. The appropriate ionization conditions for different enzymes are also differ but generally ranges at pH 6-8.14 The enzyme can be denatured due to changes in pH. The enzyme works at neutral pH and will become inactive when the environment becomes very acidic or very alkaline.15 Based on the growth ability in that pH range, Bacillus sp, and A. hydrophila can be classified into the neutrophils bacteria. Neutrophil is a bacterial groups were able to grow at pH 6-8.14,15 The growth of Bacillus sp andAeromonas hydrophila in NB medium containing 10 ppm of HgCl2 and incubated for 3 days has not reached the stationary phase. The results obtained were different with control Bacillus sp, which reached stationary phase at 37th and A. hydrophila which reached the stationary phase at the 44th and death phase in the 68th hour (Figure 7). During the period of incubation with medium containing 10 ppm HgCl2, both of these bacteria were only able to reach the exponential phase. Bacillus sp achieve exponential phase at 68th and A. hydrophila at 48th hours (Figure 7). This is because the adaptation phase is long enough. This is due to HgCl2in the medium.In the adaptation phase the synthesis of the new enzymes occurs, according to the media and the increase of cell numbers not found.14 The length of the adaptation phase in medium containing HgCl2 occur in bacteria Ochrobactrum sp S79 and L6T2 isolates, wherein the second stationary phase of these bacteria occurs on day 4 to day 9 of incubation time.16
|
|
Figure 7: The growth curveof bacteriain |
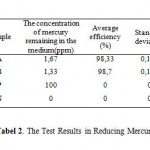
Aeromonash ydrophila and Bacilluss presistantandable to reduce mercury levels of 100 ppm to 1.67ppm for A.hydro philaand 1.33ppm for Bacilluss pafterincubated for 7days. The results of mercury content remainingin the medium, were used to determine the efficiency of both bacteriain reducing mercury. Bacillusspable to reduce mercury by 98.7%, where as A. hydro phila was 98.33% (Tabel 2). The ability of Bacillus spand A.hydrophilain reducing mercury levels associated with acharacter that is resistant tomercury. Bacterial resistance to mercury due to themer operoncontained in the plasmid.4,5
|
|
Table 2: The Test Results in Reducing Mercury |
Mer operon consists of a wide variety of mergenes. Each bacteriumhasits own mergene variations in the mer operon.3 But the mechanism of bacterial resistance to inorganic mercuryis almost the samein different bacteria species. This is due to the reduction of mercury from Hg2+ to Hg0 induced by mercuric ion reducta seen zymeen coded by the mer operon genes MerA.2 Mercuric ion reductase forms a bond with Hg2+and reduction occurs by the transfer of electrons through the flav in bond from NADPH in to NADP, so that reduced Hg was formed, ieHg0.5 The reduction of Hg2+ to Hg0is away to remove oxidized mercury and to reduce mercury dissolved in a medium.17 Some bacteria of the Genus Bacillus are known to have a gene variationin meroperon. Bacillusmegaterium and Bacillusmacroidesis abroad-spectrum mercury-resistant bacteria, where as Bacillus cereus and Bacilluslicheniformisare an arrow-spectrum mercury-resistant bacteria.18 Bacteria which only has mercury reducta seprotein(MerA) is called by a narrow spectrum mercury-resistant bacteria, while broad-spectrum mercury-resistant bacteria are bacteria that have mercury reduct as eprote in (MerA) and protein organ omerkurily ase (MerB). MerB functionsin catalyzing thetermination of the mercury-carbon bond to produce organic compound sandionic Hgin the form of saltthiols.20 Bacilluss pand A. hydrophila found in this study are not known the extent of the spectrum which is owned in reducing mercury. Until now there has been no reports of mergene variations that are owned by Aeromonas hydrophila. Howeverother species of the Genus Aeromonasare known variations in the mer operon genes. Aeromonassalmonicidah as some mergenes in the mer operon, namely Me r A, Mer P, MerR, MerE, MerT, MerD, and MerB.19 Aeromonas hydro philais able to change the shape of the cells,from rod into a round shape after mercury exposure.11
Acknowledgement
The authors would like to express their aprreciation to the Head of Magister Biology Science and Udayana University Bali Indonesia for supporting this study. Appreciation is also send toGovernor of Maluku province, Indonesia for the supportin carrying out this research work.
References
2. UNEP. Global Mercury Assesment. InterOrganization Programme for The Sound Management of Chemicals.Issued by UNEP Chemicals. Geneva, Switzerland. (2002)
3. Okoronkwo, N. E., Igwe, J. C., Okoronkwo, I. J.African Journal of Biotechnology 6(4): 337 (2006)
4. Dash, H.R., Das, S. International Biodeterioration and Biodegradation. 75 : 207-213 (2012).
5. Barkay, T., Susan, M. M., Anne, O. S. FEMS Microbiology Reviews. 27: 355-384 (2003).
6. Dubey, R.C., D.K. Maheshwari. Practical Microbiology. S.Chand and Company LTD, New Delhi, 37 (2007)
7. Osborn, A.M., Kenneth, D.B., Peter, S., Donald, A.R. FEMS Microbiology Reviews. 19: 239-262 (1997)
8. Manampiring, A.E., Billy, J.K. Jurnal Ilmiah Sains. 11 (1): 26-30 (2011)
9. Zulaika, E., Langkah, S., Agus, S. Journal of Basic and Applied Scientific Research, 2(7): 7263-7269 (2012)
10. Chaerun, S.K., Sakinah, H., Edy, S., Maelita, R.M. Microbiology, 6(2): 57-68 (2012)
11. Saidi, N., Rihab, L., Fethi, B.A., Karima, B.R., Amina, B. African Journal of Microbiology Research 7(50): 5697-5708 (2013)
12. Pelczar, M.J., Chan, E.C.S. Basic of Microbiology. Universitas Indonesia Publisher, Jakarta :138-139 (2010)
13. Perry, J.J., James, T.S., Stephen, L. 2002. Microbial Life. Sinauer Associates Publishers,Massachusetts: 142: (2002)
14. Purwoko, T. 2009. Physiology of Microbe. Penerbit Bumi Aksara, Jakarta, 234 (2009)
15. McKane, L., Judy, K. 1996. Microbiology Essentials and Aplications Second Edition. McGraw-Hill, USA, (1996)
16. Imamuddin, H. Jurnal Ekosains 2(1); 26-32 (2010).
17. Chowdury, S., Bala, N.N., Dhauria, P. 2012. International Journal of Pharmaceutical, Chemical, and Biological Science. 2(4), 600- 611 (2011).
18. Narita, M., Kazuyuki, C., Hiroshi, N., Hidenori, I., Chieh-Chen, H., Zen’ichiro, K., Simon, S., Ginro, E. FEMS Microbiology Reviews. 223; 73-82 (2003)
19. McIntosh, D., Michelle, C., Baijing, J., Frank, A.F., Erin, M.P., Sarah, E.C., Zachary, B.Z., Ilana, C.G., Russel, D., Keith, A.J., Mike, B., Rachael, R. Journal of Antimicrobial Chemotheraphy. 61; 1221-1228 (2008)
20. Huang, C.C., Chien, M.F., Lin, K.H. Interdisciplinary Studies on Environmental Chemistry — Biological Responses to ContaminantsEds., 23-29 (2010).